Details of the Target
General Information of Target
| Target ID | LDTP10563 | |||||
|---|---|---|---|---|---|---|
| Target Name | Caspase recruitment domain-containing protein 19 (CARD19) | |||||
| Gene Name | CARD19 | |||||
| Gene ID | 84270 | |||||
| Synonyms |
C9orf89; Caspase recruitment domain-containing protein 19; Bcl10-interacting CARD protein; BinCARD |
|||||
| 3D Structure | ||||||
| Sequence |
MNRKWEAKLKQIEERASHYERKPLSSVYRPRLSKPEEPPSIWRLFHRQAQAFNFVKSCKE
DVHVFALECKVGDGQRIYLVTTYAEFWFYYKSRKNLLHCYEVIPENAVCKLYFDLEFNKP ANPGADGKKMVALLIEYVCKALQELYGVNCSAEDVLNLDSSTDEKFSRHLIFQLHDVAFK DNIHVGNFLRKILQPALDLLGSEDDDSAPETTGHGFPHFSEAPARQGFSFNKMFTEKATE ESWTSNSKKLERLGSAEQSSPDLSFLVVKNNMGEKHLFVDLGVYTRNRNFRLYKSSKIGK RVALEVTEDNKFFPIQSKDVSDEYQYFLSSLVSNVRFSDTLRILTCEPSQNKQKGVGYFN SIGTSVETIEGFQCSPYPEVDHFVLSLVNKDGIKGGIRRWNYFFPEELLVYDICKYRWCE NIGRAHKSNNIMILVDLKNEVWYQKCHDPVCKAENFKSDCFPLPAEVCLLFLFKEEEEFT TDEADETRSNETQNPHKPSPSRLSTGASADAVWDNGIDDAYFLEATEDAELAEAAENSLL SYNSEVDEIPDELIIEVLQE |
|||||
| Target Bioclass |
Other
|
|||||
| Subcellular location |
Nucleus; Endoplasmic reticulum membrane
|
|||||
| Function | Plays a role in inhibiting the effects of BCL10-induced activation of NF-kappa-B. May inhibit the phosphorylation of BCL10 in a CARD-dependent manner. | |||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
14.71 | LDD0402 | [1] | |
|
Jackson_1 Probe Info |
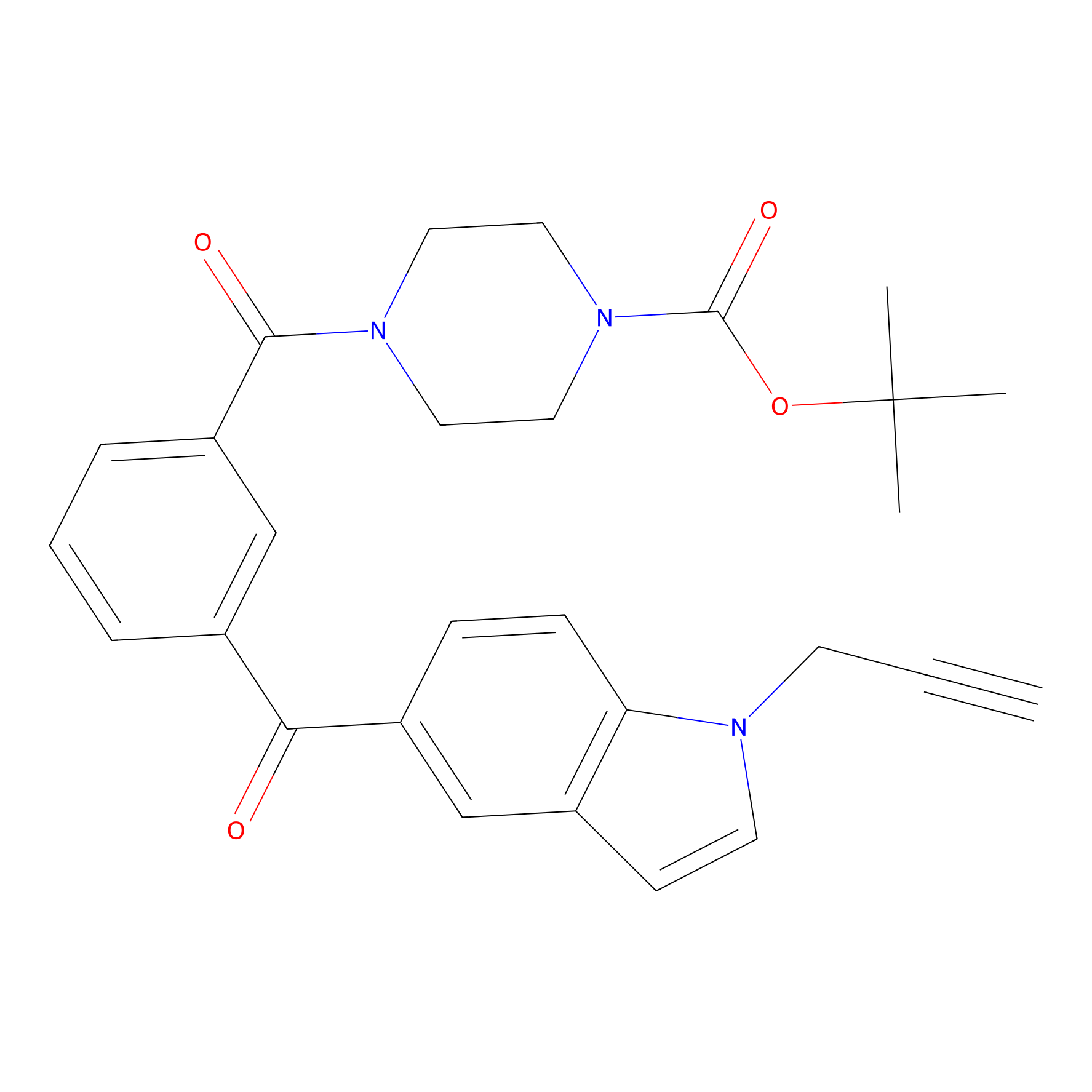 |
20.00 | LDD0122 | [2] | |
|
TH211 Probe Info |
 |
Y85(11.08) | LDD0260 | [3] | |
|
DBIA Probe Info |
 |
C63(1.60) | LDD3310 | [4] | |
|
AHL-Pu-1 Probe Info |
 |
C63(2.79) | LDD0171 | [5] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0227 | [6] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0165 | [7] | |
|
IPM Probe Info |
 |
N.A. | LDD0005 | [8] | |
|
TFBX Probe Info |
 |
N.A. | LDD0148 | [9] | |
|
NAIA_5 Probe Info |
 |
C63(0.00); C7(0.00) | LDD2223 | [10] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0025 | 4SU-RNA | HEK-293T | C63(2.10) | LDD0371 | [5] |
| LDCM0026 | 4SU-RNA+native RNA | DM93 | C63(2.79) | LDD0171 | [5] |
| LDCM0632 | CL-Sc | Hep-G2 | C63(0.90) | LDD2227 | [10] |
| LDCM0213 | Electrophilic fragment 2 | MDA-MB-231 | C63(1.66) | LDD1702 | [11] |
| LDCM0572 | Fragment10 | Ramos | C63(0.98) | LDD2189 | [12] |
| LDCM0573 | Fragment11 | Ramos | C63(20.00) | LDD2190 | [12] |
| LDCM0574 | Fragment12 | Ramos | C63(0.75) | LDD2191 | [12] |
| LDCM0575 | Fragment13 | Ramos | C63(0.94) | LDD2192 | [12] |
| LDCM0576 | Fragment14 | Ramos | C63(0.65) | LDD2193 | [12] |
| LDCM0579 | Fragment20 | Ramos | C63(0.82) | LDD2194 | [12] |
| LDCM0580 | Fragment21 | Ramos | C63(0.99) | LDD2195 | [12] |
| LDCM0582 | Fragment23 | Ramos | C63(1.09) | LDD2196 | [12] |
| LDCM0578 | Fragment27 | Ramos | C63(0.77) | LDD2197 | [12] |
| LDCM0586 | Fragment28 | Ramos | C63(0.84) | LDD2198 | [12] |
| LDCM0588 | Fragment30 | Ramos | C63(0.84) | LDD2199 | [12] |
| LDCM0589 | Fragment31 | Ramos | C63(1.01) | LDD2200 | [12] |
| LDCM0590 | Fragment32 | Ramos | C63(0.65) | LDD2201 | [12] |
| LDCM0468 | Fragment33 | Ramos | C63(0.88) | LDD2202 | [12] |
| LDCM0596 | Fragment38 | Ramos | C63(0.94) | LDD2203 | [12] |
| LDCM0614 | Fragment56 | Ramos | C63(1.22) | LDD2205 | [12] |
| LDCM0571 | Fragment9 | Ramos | C63(0.99) | LDD2188 | [12] |
| LDCM0022 | KB02 | 769-P | C63(1.67) | LDD2246 | [4] |
| LDCM0023 | KB03 | MDA-MB-231 | C63(2.80) | LDD1701 | [11] |
| LDCM0024 | KB05 | COLO792 | C63(1.60) | LDD3310 | [4] |
| LDCM0109 | NEM | HeLa | N.A. | LDD0227 | [6] |
References
