Details of the Target
General Information of Target
| Target ID | LDTP05568 | |||||
|---|---|---|---|---|---|---|
| Target Name | Pseudouridine-5'-phosphatase (PUDP) | |||||
| Gene Name | PUDP | |||||
| Gene ID | 8226 | |||||
| Synonyms |
DXF68S1E; FAM16AX; GS1; HDHD1; HDHD1A; Pseudouridine-5'-phosphatase; EC 3.1.3.96; Haloacid dehalogenase-like hydrolase domain-containing protein 1; Haloacid dehalogenase-like hydrolase domain-containing protein 1A; Protein GS1; Pseudouridine-5'-monophosphatase; 5'-PsiMPase
|
|||||
| 3D Structure | ||||||
| Sequence |
MAAPPQPVTHLIFDMDGLLLDTERLYSVVFQEICNRYDKKYSWDVKSLVMGKKALEAAQI
IIDVLQLPMSKEELVEESQTKLKEVFPTAALMPGAEKLIIHLRKHGIPFALATSSGSASF DMKTSRHKEFFSLFSHIVLGDDPEVQHGKPDPDIFLACAKRFSPPPAMEKCLVFEDAPNG VEAALAAGMQVVMVPDGNLSRDLTTKATLVLNSLQDFQPELFGLPSYE |
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
HAD-like hydrolase superfamily, CbbY/CbbZ/Gph/YieH family
|
|||||
| Function | Dephosphorylates pseudouridine 5'-phosphate, a potential intermediate in rRNA degradation. Pseudouridine is then excreted intact in urine. | |||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
STPyne Probe Info |
 |
K46(5.79); K83(8.26) | LDD0277 | [1] | |
|
HHS-475 Probe Info |
 |
Y26(1.15) | LDD0264 | [2] | |
|
HHS-465 Probe Info |
 |
Y26(10.00) | LDD2237 | [3] | |
|
DBIA Probe Info |
 |
C171(1.13) | LDD1508 | [4] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
N.A. | LDD0038 | [5] | |
|
IA-alkyne Probe Info |
 |
C34(0.00); C171(0.00) | LDD0036 | [5] | |
|
Lodoacetamide azide Probe Info |
 |
C34(0.00); C171(0.00) | LDD0037 | [5] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0217 | [6] | |
|
NAIA_5 Probe Info |
 |
C171(0.00); C158(0.00); C34(0.00) | LDD2223 | [7] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C286 Probe Info |
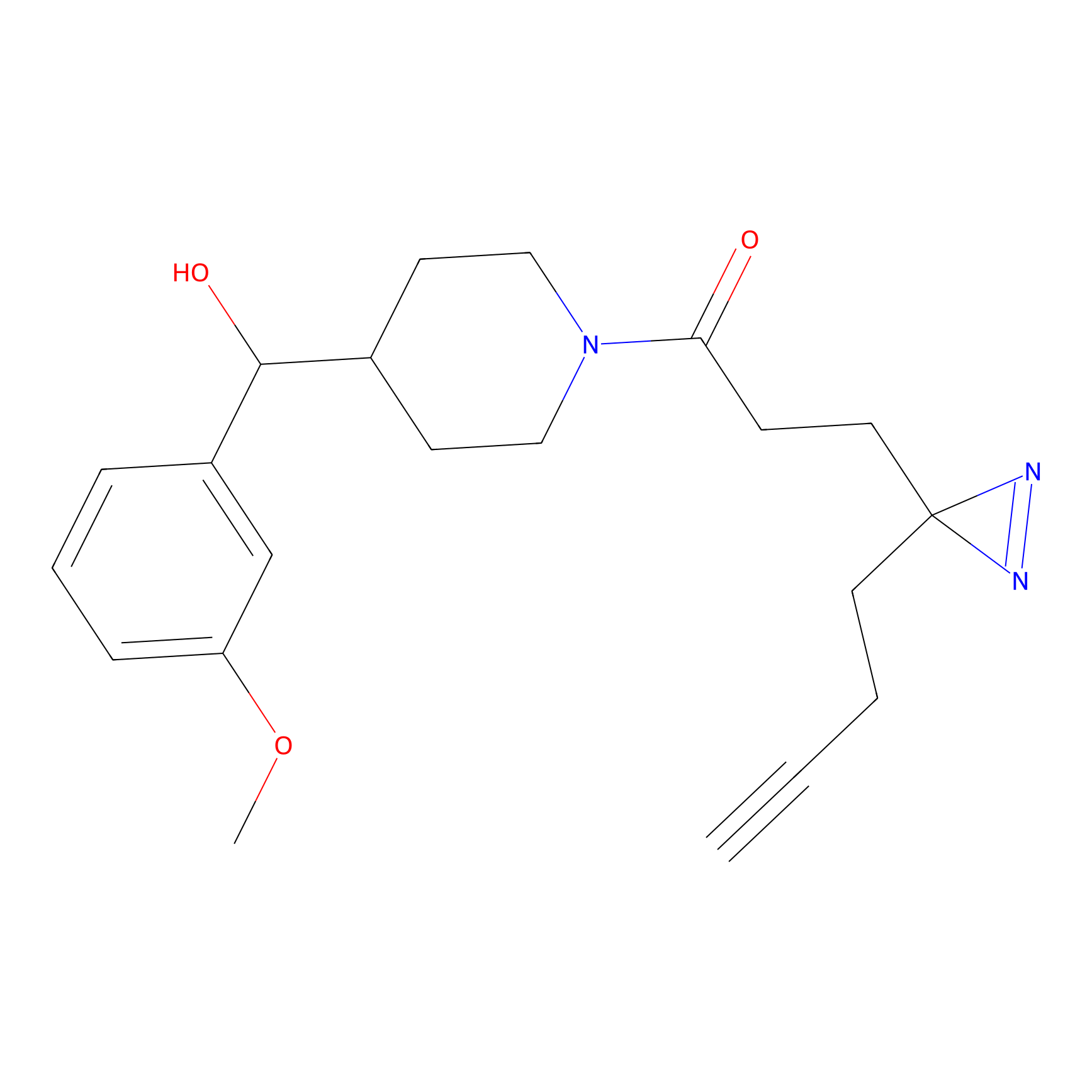 |
7.46 | LDD1956 | [8] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0215 | AC10 | HEK-293T | C171(1.13) | LDD1508 | [4] |
| LDCM0277 | AC18 | HEK-293T | C171(1.00) | LDD1516 | [4] |
| LDCM0279 | AC2 | HEK-293T | C171(1.14) | LDD1518 | [4] |
| LDCM0286 | AC26 | HEK-293T | C171(1.30) | LDD1525 | [4] |
| LDCM0295 | AC34 | HEK-293T | C171(0.92) | LDD1534 | [4] |
| LDCM0304 | AC42 | HEK-293T | C171(0.87) | LDD1543 | [4] |
| LDCM0313 | AC50 | HEK-293T | C171(1.14) | LDD1552 | [4] |
| LDCM0321 | AC58 | HEK-293T | C171(1.13) | LDD1560 | [4] |
| LDCM0108 | Chloroacetamide | HeLa | N.A. | LDD0222 | [6] |
| LDCM0369 | CL100 | HEK-293T | C171(0.93) | LDD1573 | [4] |
| LDCM0373 | CL104 | HEK-293T | C171(1.11) | LDD1577 | [4] |
| LDCM0377 | CL108 | HEK-293T | C171(1.30) | LDD1581 | [4] |
| LDCM0382 | CL112 | HEK-293T | C171(0.94) | LDD1586 | [4] |
| LDCM0386 | CL116 | HEK-293T | C171(1.06) | LDD1590 | [4] |
| LDCM0391 | CL120 | HEK-293T | C171(1.30) | LDD1595 | [4] |
| LDCM0395 | CL124 | HEK-293T | C171(0.96) | LDD1599 | [4] |
| LDCM0399 | CL128 | HEK-293T | C171(1.06) | LDD1603 | [4] |
| LDCM0403 | CL16 | HEK-293T | C171(1.31) | LDD1607 | [4] |
| LDCM0405 | CL18 | HEK-293T | C171(1.01) | LDD1609 | [4] |
| LDCM0416 | CL28 | HEK-293T | C171(0.94) | LDD1620 | [4] |
| LDCM0419 | CL30 | HEK-293T | C171(1.22) | LDD1623 | [4] |
| LDCM0429 | CL4 | HEK-293T | C171(1.07) | LDD1633 | [4] |
| LDCM0430 | CL40 | HEK-293T | C171(0.92) | LDD1634 | [4] |
| LDCM0432 | CL42 | HEK-293T | C171(1.02) | LDD1636 | [4] |
| LDCM0443 | CL52 | HEK-293T | C171(1.29) | LDD1646 | [4] |
| LDCM0445 | CL54 | HEK-293T | C171(1.18) | LDD1648 | [4] |
| LDCM0451 | CL6 | HEK-293T | C171(0.94) | LDD1654 | [4] |
| LDCM0456 | CL64 | HEK-293T | C171(1.16) | LDD1659 | [4] |
| LDCM0458 | CL66 | HEK-293T | C171(1.08) | LDD1661 | [4] |
| LDCM0469 | CL76 | HEK-293T | C171(0.97) | LDD1672 | [4] |
| LDCM0471 | CL78 | HEK-293T | C171(1.13) | LDD1674 | [4] |
| LDCM0482 | CL88 | HEK-293T | C171(0.92) | LDD1685 | [4] |
| LDCM0485 | CL90 | HEK-293T | C171(0.87) | LDD1688 | [4] |
| LDCM0576 | Fragment14 | Ramos | C34(0.54) | LDD2193 | [9] |
| LDCM0566 | Fragment4 | Ramos | C34(0.80) | LDD2184 | [9] |
| LDCM0569 | Fragment7 | Ramos | C34(0.38) | LDD2186 | [9] |
| LDCM0116 | HHS-0101 | DM93 | Y26(1.15) | LDD0264 | [2] |
| LDCM0117 | HHS-0201 | DM93 | Y26(1.70) | LDD0265 | [2] |
| LDCM0118 | HHS-0301 | DM93 | Y26(1.45) | LDD0266 | [2] |
| LDCM0119 | HHS-0401 | DM93 | Y26(2.00) | LDD0267 | [2] |
| LDCM0120 | HHS-0701 | DM93 | Y26(1.29) | LDD0268 | [2] |
| LDCM0107 | IAA | HeLa | N.A. | LDD0221 | [6] |
| LDCM0022 | KB02 | Ramos | C34(0.26) | LDD2182 | [9] |
| LDCM0023 | KB03 | Ramos | C34(0.30) | LDD2183 | [9] |
| LDCM0024 | KB05 | Ramos | C34(0.24) | LDD2185 | [9] |
| LDCM0109 | NEM | HeLa | N.A. | LDD0224 | [6] |
| LDCM0131 | RA190 | MM1.R | C34(1.12) | LDD0304 | [10] |
The Interaction Atlas With This Target
References
