Details of the Target
General Information of Target
| Target ID | LDTP05010 | |||||
|---|---|---|---|---|---|---|
| Target Name | Ubiquitin-conjugating enzyme E2 D2 (UBE2D2) | |||||
| Gene Name | UBE2D2 | |||||
| Gene ID | 7322 | |||||
| Synonyms |
PUBC1; UBC4; UBC5B; UBCH4; UBCH5B; Ubiquitin-conjugating enzyme E2 D2; EC 2.3.2.23; (E3-independent) E2 ubiquitin-conjugating enzyme D2; EC 2.3.2.24; E2 ubiquitin-conjugating enzyme D2; Ubiquitin carrier protein D2; Ubiquitin-conjugating enzyme E2(17)KB 2; Ubiquitin-conjugating enzyme E2-17 kDa 2; Ubiquitin-protein ligase D2; p53-regulated ubiquitin-conjugating enzyme 1
|
|||||
| 3D Structure | ||||||
| Sequence |
MALKRIHKELNDLARDPPAQCSAGPVGDDMFHWQATIMGPNDSPYQGGVFFLTIHFPTDY
PFKPPKVAFTTRIYHPNINSNGSICLDILRSQWSPALTISKVLLSICSLLCDPNPDDPLV PEIARIYKTDREKYNRIAREWTQKYAM |
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
Ubiquitin-conjugating enzyme family
|
|||||
| Function |
Accepts ubiquitin from the E1 complex and catalyzes its covalent attachment to other proteins. Catalyzes 'Lys-48'-linked polyubiquitination. Mediates the selective degradation of short-lived and abnormal proteins. Functions in the E6/E6-AP-induced ubiquitination of p53/TP53. Mediates ubiquitination of PEX5 and SQSTM1 and autoubiquitination of STUB1 and TRAF6. Involved in the signal-induced conjugation and subsequent degradation of NFKBIA, FBXW2-mediated GCM1 ubiquitination and degradation, MDM2-dependent degradation of p53/TP53 and the activation of MAVS in the mitochondria by RIGI in response to viral infection. Essential for viral activation of IRF3.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
| ChEMBL ID | ||||||
Target Site Mutations in Different Cell Lines
| Cell line | Mutation details | Probe for labeling this protein in this cell | |||
|---|---|---|---|---|---|
| LS180 | Deletion: p.T98FfsTer21 | DBIA Probe Info | |||
| MCC26 | SNV: p.L119F | DBIA Probe Info | |||
| MFE319 | SNV: p.N81S | DBIA Probe Info | |||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
STPyne Probe Info |
 |
K128(1.41); K8(6.60) | LDD0277 | [1] | |
|
ONAyne Probe Info |
 |
N.A. | LDD0273 | [1] | |
|
DBIA Probe Info |
 |
C87(1.80) | LDD3310 | [2] | |
|
JZ128-DTB Probe Info |
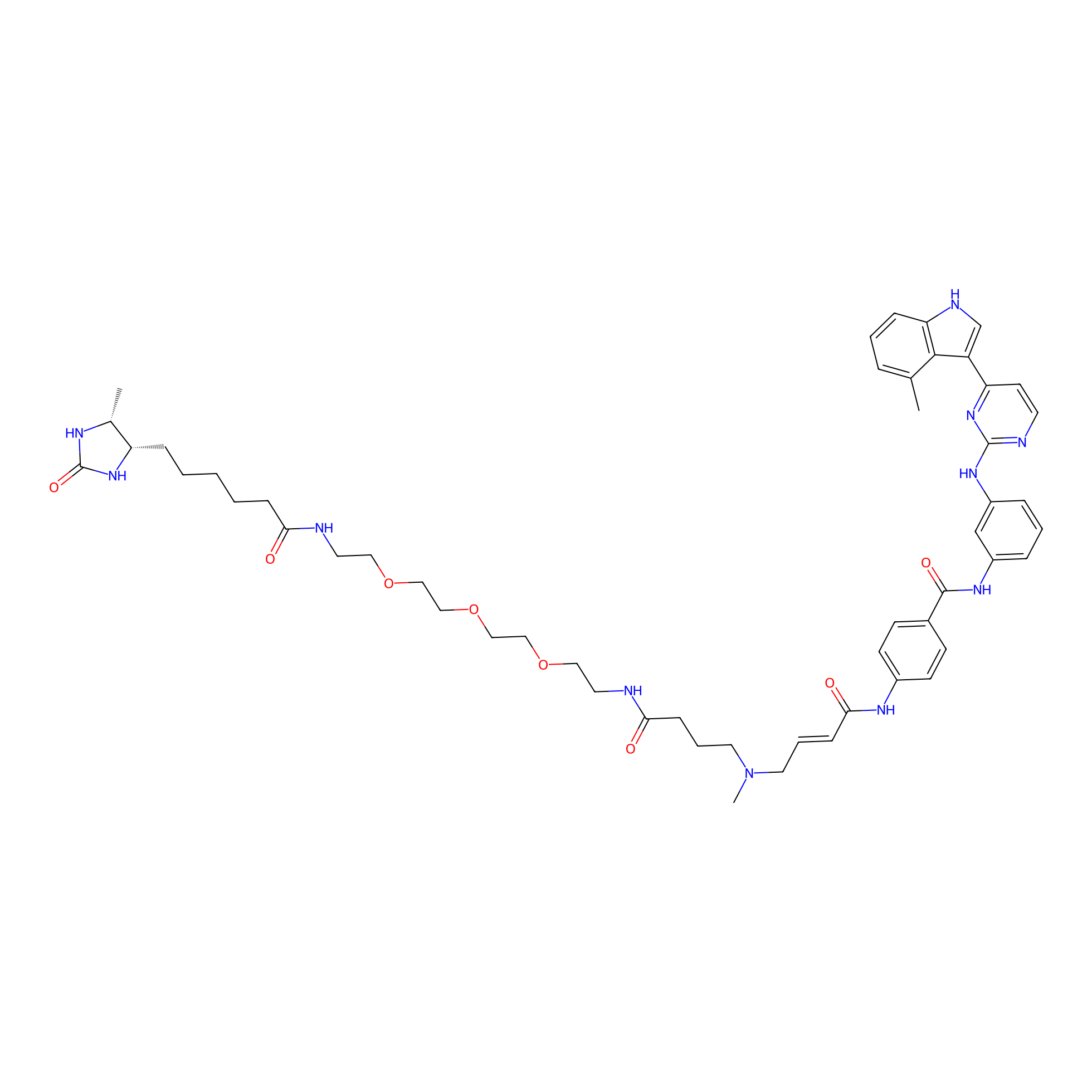 |
N.A. | LDD0462 | [3] | |
|
AHL-Pu-1 Probe Info |
 |
C107(2.63) | LDD0168 | [4] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0174 | [5] | |
|
NAIA_4 Probe Info |
 |
C85(0.00); C107(0.00); C111(0.00) | LDD2226 | [6] | |
|
NAIA_5 Probe Info |
 |
N.A. | LDD2224 | [6] | |
|
Compound 10 Probe Info |
 |
N.A. | LDD2216 | [7] | |
|
IPM Probe Info |
 |
C111(0.00); C85(0.00) | LDD2156 | [8] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0217 | [9] | |
|
AOyne Probe Info |
 |
15.00 | LDD0443 | [10] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0025 | 4SU-RNA | HEK-293T | C107(2.63) | LDD0168 | [4] |
| LDCM0026 | 4SU-RNA+native RNA | HEK-293T | C107(2.30) | LDD0169 | [4] |
| LDCM0108 | Chloroacetamide | HeLa | C85(0.00); H75(0.00) | LDD0222 | [9] |
| LDCM0107 | IAA | HeLa | N.A. | LDD0221 | [9] |
| LDCM0179 | JZ128 | PC-3 | N.A. | LDD0462 | [3] |
| LDCM0022 | KB02 | 22RV1 | C87(1.33) | LDD2243 | [2] |
| LDCM0023 | KB03 | 22RV1 | C87(1.35) | LDD2660 | [2] |
| LDCM0024 | KB05 | COLO792 | C87(1.80) | LDD3310 | [2] |
| LDCM0109 | NEM | HeLa | N.A. | LDD0223 | [9] |
| LDCM0628 | OTUB2-COV-1 | HEK-293T | C85(0.75) | LDD2207 | [11] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Transporter and channel
Other
References
