Details of the Target
General Information of Target
| Target ID | LDTP04414 | |||||
|---|---|---|---|---|---|---|
| Target Name | Tyrosine-protein kinase Blk (BLK) | |||||
| Gene Name | BLK | |||||
| Gene ID | 640 | |||||
| Synonyms |
Tyrosine-protein kinase Blk; EC 2.7.10.2; B lymphocyte kinase; p55-Blk |
|||||
| 3D Structure | ||||||
| Sequence |
MGLVSSKKPDKEKPIKEKDKGQWSPLKVSAQDKDAPPLPPLVVFNHLTPPPPDEHLDEDK
HFVVALYDYTAMNDRDLQMLKGEKLQVLKGTGDWWLARSLVTGREGYVPSNFVARVESLE MERWFFRSQGRKEAERQLLAPINKAGSFLIRESETNKGAFSLSVKDVTTQGELIKHYKIR CLDEGGYYISPRITFPSLQALVQHYSKKGDGLCQRLTLPCVRPAPQNPWAQDEWEIPRQS LRLVRKLGSGQFGEVWMGYYKNNMKVAIKTLKEGTMSPEAFLGEANVMKALQHERLVRLY AVVTKEPIYIVTEYMARGCLLDFLKTDEGSRLSLPRLIDMSAQIAEGMAYIERMNSIHRD LRAANILVSEALCCKIADFGLARIIDSEYTAQEGAKFPIKWTAPEAIHFGVFTIKADVWS FGVLLMEVVTYGRVPYPGMSNPEVIRNLERGYRMPRPDTCPPELYRGVIAECWRSRPEER PTFEFLQSVLEDFYTATERQYELQP |
|||||
| Target Type |
Patented-recorded
|
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
Protein kinase superfamily, Tyr protein kinase family, SRC subfamily
|
|||||
| Subcellular location |
Cell membrane
|
|||||
| Function |
Non-receptor tyrosine kinase involved in B-lymphocyte development, differentiation and signaling. B-cell receptor (BCR) signaling requires a tight regulation of several protein tyrosine kinases and phosphatases, and associated coreceptors. Binding of antigen to the B-cell antigen receptor (BCR) triggers signaling that ultimately leads to B-cell activation. Signaling through BLK plays an important role in transmitting signals through surface immunoglobulins and supports the pro-B to pre-B transition, as well as the signaling for growth arrest and apoptosis downstream of B-cell receptor. Specifically binds and phosphorylates CD79A at 'Tyr-188'and 'Tyr-199', as well as CD79B at 'Tyr-196' and 'Tyr-207'. Phosphorylates also the immunoglobulin G receptors FCGR2A, FCGR2B and FCGR2C. With FYN and LYN, plays an essential role in pre-B-cell receptor (pre-BCR)-mediated NF-kappa-B activation. Contributes also to BTK activation by indirectly stimulating BTK intramolecular autophosphorylation. In pancreatic islets, acts as a modulator of beta-cells function through the up-regulation of PDX1 and NKX6-1 and consequent stimulation of insulin secretion in response to glucose. Phosphorylates CGAS, promoting retention of CGAS in the cytosol.
|
|||||
| TTD ID | ||||||
| Uniprot ID | ||||||
| DrugMap ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
| ChEMBL ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
IBr-1b Probe Info |
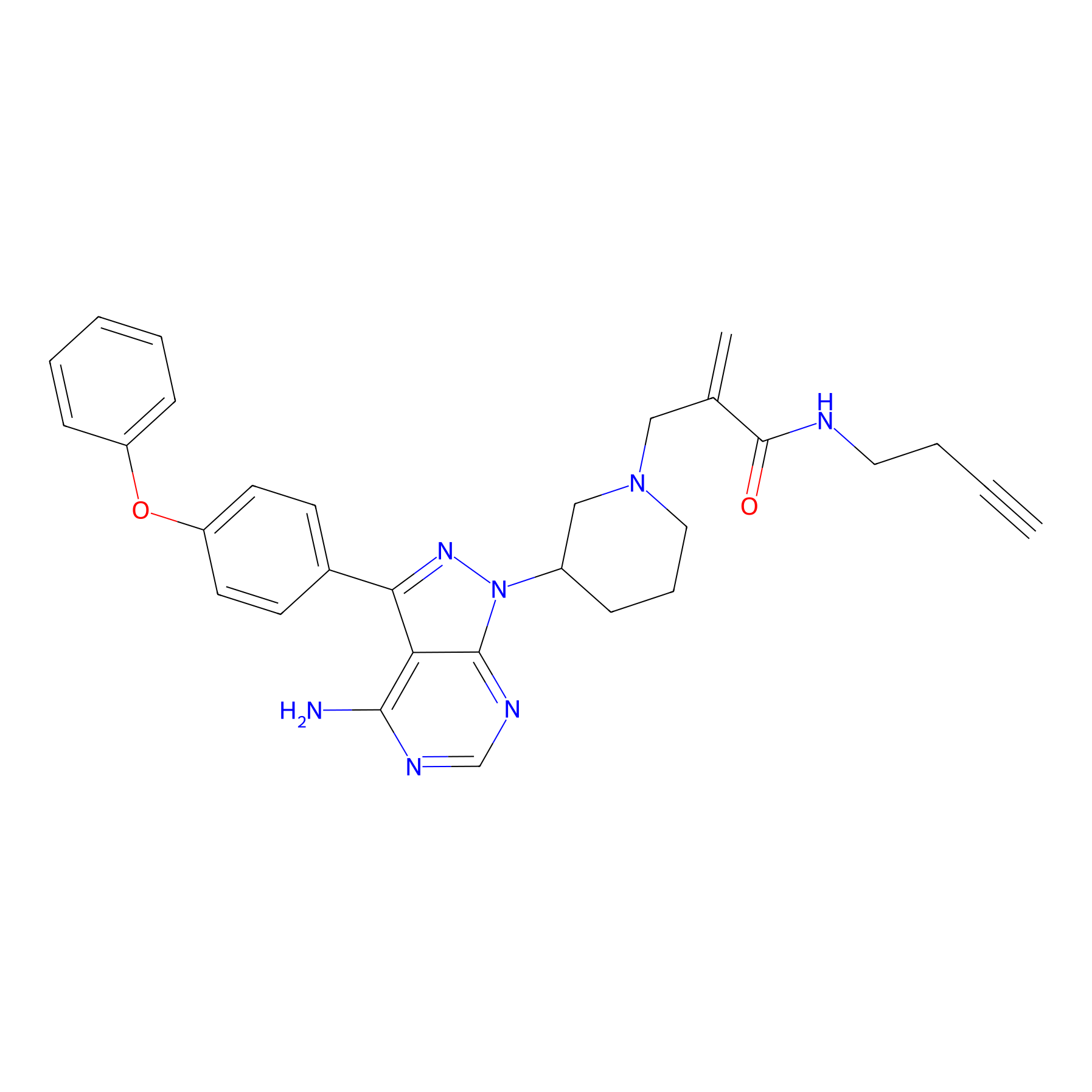 |
20.00 | LDD0306 | [1] | |
|
DBIA Probe Info |
 |
C319(17.88) | LDD0204 | [2] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0231 | [3] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0036 | [4] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
STS-1 Probe Info |
 |
1.17 | LDD0137 | [5] | |
|
STS-2 Probe Info |
 |
1.00 | LDD0139 | [5] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0102 | BDHI 8 | Jurkat | C319(17.88) | LDD0204 | [2] |
| LDCM0206 | EV-99 | T cell | C319(20.00) | LDD0521 | [6] |
| LDCM0625 | F8 | Ramos | C181(1.12); C319(0.32); C472(1.59); C213(1.38) | LDD2187 | [7] |
| LDCM0572 | Fragment10 | Ramos | C181(1.07); C319(1.61); C472(1.78); C213(1.74) | LDD2189 | [7] |
| LDCM0573 | Fragment11 | Ramos | C181(0.69); C319(3.94); C472(0.39); C213(0.93) | LDD2190 | [7] |
| LDCM0574 | Fragment12 | Ramos | C181(1.20); C319(3.76); C472(0.77); C213(1.51) | LDD2191 | [7] |
| LDCM0575 | Fragment13 | Ramos | C181(0.99); C319(2.25); C213(0.80); C220(0.32) | LDD2192 | [7] |
| LDCM0576 | Fragment14 | Ramos | C181(1.17); C472(0.97); C213(1.93); 1.44 | LDD2193 | [7] |
| LDCM0579 | Fragment20 | Ramos | C181(1.28); C319(4.35); C472(1.58); C460(0.59) | LDD2194 | [7] |
| LDCM0580 | Fragment21 | Ramos | C181(1.03); C319(0.95); C472(0.76); C213(0.82) | LDD2195 | [7] |
| LDCM0582 | Fragment23 | Ramos | C181(1.09); C319(0.63); C472(0.85); C213(1.19) | LDD2196 | [7] |
| LDCM0578 | Fragment27 | Ramos | C181(0.98); C319(0.85); C472(0.95); C213(0.91) | LDD2197 | [7] |
| LDCM0586 | Fragment28 | Ramos | C181(0.82); C319(0.94); C472(0.62); C213(0.43) | LDD2198 | [7] |
| LDCM0588 | Fragment30 | Ramos | C181(1.01); C319(20.00); C472(0.78); C213(1.04) | LDD2199 | [7] |
| LDCM0589 | Fragment31 | Ramos | C181(1.02); C319(0.84); C472(0.90); C213(0.84) | LDD2200 | [7] |
| LDCM0590 | Fragment32 | Ramos | C181(1.03); C319(6.73); C472(0.58); C460(1.20) | LDD2201 | [7] |
| LDCM0468 | Fragment33 | Ramos | C181(0.80); C319(1.05); C472(0.69); C213(0.73) | LDD2202 | [7] |
| LDCM0596 | Fragment38 | Ramos | C181(0.77); C319(0.80); C472(0.55); C213(1.38) | LDD2203 | [7] |
| LDCM0566 | Fragment4 | Ramos | C181(1.14); C319(1.53); C472(2.33); C213(1.70) | LDD2184 | [7] |
| LDCM0610 | Fragment52 | Ramos | C181(0.98); C472(0.76); C213(1.21); C220(0.37) | LDD2204 | [7] |
| LDCM0614 | Fragment56 | Ramos | C181(1.01); C319(0.94); C472(0.72); C213(1.21) | LDD2205 | [7] |
| LDCM0569 | Fragment7 | Ramos | C181(1.50); C319(7.56); C472(1.44); C213(2.19) | LDD2186 | [7] |
| LDCM0571 | Fragment9 | Ramos | C181(1.11); C319(2.42); C472(3.61); C460(6.18) | LDD2188 | [7] |
| LDCM0022 | KB02 | Ramos | C181(1.24); C319(4.79); C472(1.17); C213(2.17) | LDD2182 | [7] |
| LDCM0023 | KB03 | Jurkat | C319(69.57) | LDD0209 | [2] |
| LDCM0024 | KB05 | NALM-6 | C181(2.13); C319(4.39); C472(1.41); C220(1.28) | LDD3339 | [8] |
| LDCM0109 | NEM | HeLa | N.A. | LDD0231 | [3] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Heat shock protein HSP 90-beta (HSP90AB1) | Heat shock protein 90 family | P08238 | |||
Transcription factor
Other
The Drug(s) Related To This Target
Approved
| Drug Name | Drug Type | External ID | |||
|---|---|---|---|---|---|
| Fostamatinib | Small molecular drug | DB12010 | |||
| Zanubrutinib | . | DB15035 | |||
Investigative
| Drug Name | Drug Type | External ID | |||
|---|---|---|---|---|---|
| Pmid15546730c2 | Small molecular drug | D08RZB | |||
| Pmid24915291c31 | Small molecular drug | D01RHR | |||
| Pmid24915291c38 | Small molecular drug | D06BJG | |||
Patented
| Drug Name | Drug Type | External ID | |||
|---|---|---|---|---|---|
| Pyrazolo[4,3-c]Pyridine Derivative 2 | . | D09PIS | |||
References
