Details of the Target
General Information of Target
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
STPyne Probe Info |
 |
K101(2.52); K31(10.00) | LDD0277 | [1] | |
|
DBIA Probe Info |
 |
C17(3.17) | LDD3333 | [2] | |
|
Cep-alkyne Probe Info |
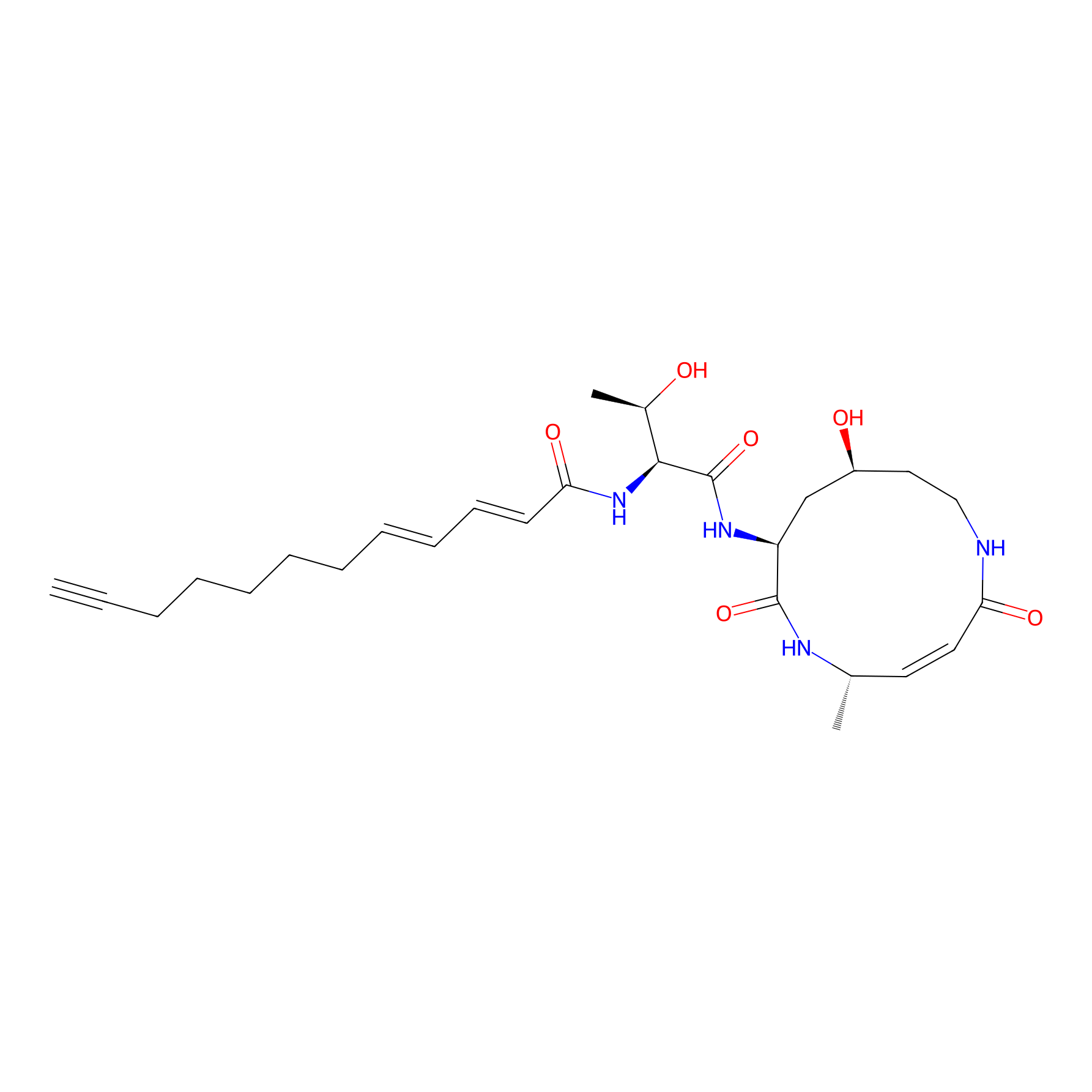 |
3.27 | LDD0284 | [3] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
N.A. | LDD0038 | [4] | |
|
IA-alkyne Probe Info |
 |
C17(0.00); C70(0.00) | LDD0036 | [4] | |
|
Lodoacetamide azide Probe Info |
 |
C17(0.00); C70(0.00) | LDD0037 | [4] | |
|
Compound 10 Probe Info |
 |
N.A. | LDD2216 | [5] | |
|
Compound 11 Probe Info |
 |
N.A. | LDD2213 | [5] | |
|
IPM Probe Info |
 |
N.A. | LDD0147 | [6] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0122 | Cepafungin 1 | RPMI-8226 | 3.27 | LDD0284 | [3] |
| LDCM0625 | F8 | Ramos | C17(0.77); C70(1.49) | LDD2187 | [7] |
| LDCM0572 | Fragment10 | Ramos | C17(1.28); C70(0.64) | LDD2189 | [7] |
| LDCM0573 | Fragment11 | Ramos | C17(0.42) | LDD2190 | [7] |
| LDCM0574 | Fragment12 | Ramos | C17(1.54); C70(0.56) | LDD2191 | [7] |
| LDCM0575 | Fragment13 | Ramos | C17(1.00); C70(0.96) | LDD2192 | [7] |
| LDCM0576 | Fragment14 | Ramos | C17(0.94); C70(0.97) | LDD2193 | [7] |
| LDCM0579 | Fragment20 | Ramos | C17(2.46); C70(0.47) | LDD2194 | [7] |
| LDCM0580 | Fragment21 | Ramos | C17(1.31); C70(0.83) | LDD2195 | [7] |
| LDCM0582 | Fragment23 | Ramos | C17(0.93); C70(0.89) | LDD2196 | [7] |
| LDCM0578 | Fragment27 | Ramos | C17(1.06); C70(0.92) | LDD2197 | [7] |
| LDCM0586 | Fragment28 | Ramos | C17(0.95); C70(0.52) | LDD2198 | [7] |
| LDCM0588 | Fragment30 | Ramos | C17(1.26); C70(1.14) | LDD2199 | [7] |
| LDCM0589 | Fragment31 | Ramos | C17(1.00); C70(0.81) | LDD2200 | [7] |
| LDCM0590 | Fragment32 | Ramos | C17(2.18); C70(0.78) | LDD2201 | [7] |
| LDCM0468 | Fragment33 | Ramos | C17(1.36); C70(0.90) | LDD2202 | [7] |
| LDCM0596 | Fragment38 | Ramos | C17(1.07); C70(0.70) | LDD2203 | [7] |
| LDCM0566 | Fragment4 | Ramos | C17(1.29); C70(0.82) | LDD2184 | [7] |
| LDCM0610 | Fragment52 | Ramos | C17(1.20); C70(1.26) | LDD2204 | [7] |
| LDCM0614 | Fragment56 | Ramos | C17(2.49); C70(1.16) | LDD2205 | [7] |
| LDCM0569 | Fragment7 | Ramos | C17(1.28); C70(0.71) | LDD2186 | [7] |
| LDCM0571 | Fragment9 | Ramos | C17(2.31); C70(0.67) | LDD2188 | [7] |
| LDCM0022 | KB02 | T cell-activated | C17(5.54) | LDD1704 | [8] |
| LDCM0023 | KB03 | Ramos | C17(1.37); C70(0.79) | LDD2183 | [7] |
| LDCM0024 | KB05 | MOLM-13 | C17(3.17) | LDD3333 | [2] |
| LDCM0131 | RA190 | MM1.R | C17(2.29) | LDD0304 | [9] |
| LDCM0170 | Structure8 | Ramos | 5.01 | LDD0433 | [10] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Proteasome subunit beta type-3 (PSMB3) | Peptidase T1B family | P49720 | |||
Transcription factor
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Cone-rod homeobox protein (CRX) | Paired homeobox family | O43186 | |||
| Homeobox protein MOX-2 (MEOX2) | . | P50222 | |||
Other
The Drug(s) Related To This Target
Approved
| Drug Name | Drug Type | External ID | |||
|---|---|---|---|---|---|
| Carfilzomib | Small molecular drug | DB08889 | |||
Patented
References
