Details of the Target
General Information of Target
| Target ID | LDTP14429 | |||||
|---|---|---|---|---|---|---|
| Target Name | Eukaryotic translation initiation factor 1A, Y-chromosomal (EIF1AY) | |||||
| Gene Name | EIF1AY | |||||
| Gene ID | 9086 | |||||
| Synonyms |
Eukaryotic translation initiation factor 1A, Y-chromosomal; eIF-1A Y isoform; eIF1A Y isoform; Eukaryotic translation initiation factor 4C; eIF-4C |
|||||
| 3D Structure | ||||||
| Sequence |
MMTLVPRARTRAGQDHYSHPCPRFSQVLLTEGIMTYCLTKNLSDVNILHRLLKNGNVRNT
LLQSKVGLLTYYVKLYPGEVTLLTRPSIQMRLCCITGSVSRPRSQK |
|||||
| Target Bioclass |
Other
|
|||||
| Family |
EIF-1A family
|
|||||
| Function |
Component of the 43S pre-initiation complex (43S PIC), which binds to the mRNA cap-proximal region, scans mRNA 5'-untranslated region, and locates the initiation codon. This protein enhances formation of the cap-proximal complex. Together with EIF1, facilitates scanning, start codon recognition, promotion of the assembly of 48S complex at the initiation codon (43S PIC becomes 48S PIC after the start codon is reached), and dissociation of aberrant complexes. After start codon location, together with EIF5B orients the initiator methionine-tRNA in a conformation that allows 60S ribosomal subunit joining to form the 80S initiation complex. Is released after 80S initiation complex formation, just after GTP hydrolysis by EIF5B, and before release of EIF5B. Its globular part is located in the A site of the 40S ribosomal subunit. Its interaction with EIF5 during scanning contribute to the maintenance of EIF1 within the open 43S PIC. In contrast to yeast orthologs, does not bind EIF1.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
STPyne Probe Info |
 |
K104(6.69); K88(3.17) | LDD0277 | [1] | |
|
AZ-9 Probe Info |
 |
E111(1.26) | LDD2208 | [2] | |
|
ONAyne Probe Info |
 |
N.A. | LDD0273 | [1] | |
|
Probe 1 Probe Info |
 |
Y35(52.51); Y84(251.24); Y95(35.00); Y106(158.96) | LDD3495 | [3] | |
|
HHS-475 Probe Info |
 |
Y106(0.82) | LDD0264 | [4] | |
|
HHS-465 Probe Info |
 |
Y106(7.24) | LDD2237 | [5] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0221 | [6] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
N.A. | LDD0038 | [7] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0036 | [7] | |
|
Lodoacetamide azide Probe Info |
 |
N.A. | LDD0037 | [7] | |
|
1c-yne Probe Info |
 |
K104(0.00); K29(0.00) | LDD0228 | [8] | |
|
Crotonaldehyde Probe Info |
 |
N.A. | LDD0219 | [6] | |
|
NAIA_5 Probe Info |
 |
N.A. | LDD2223 | [9] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
pLen Probe Info |
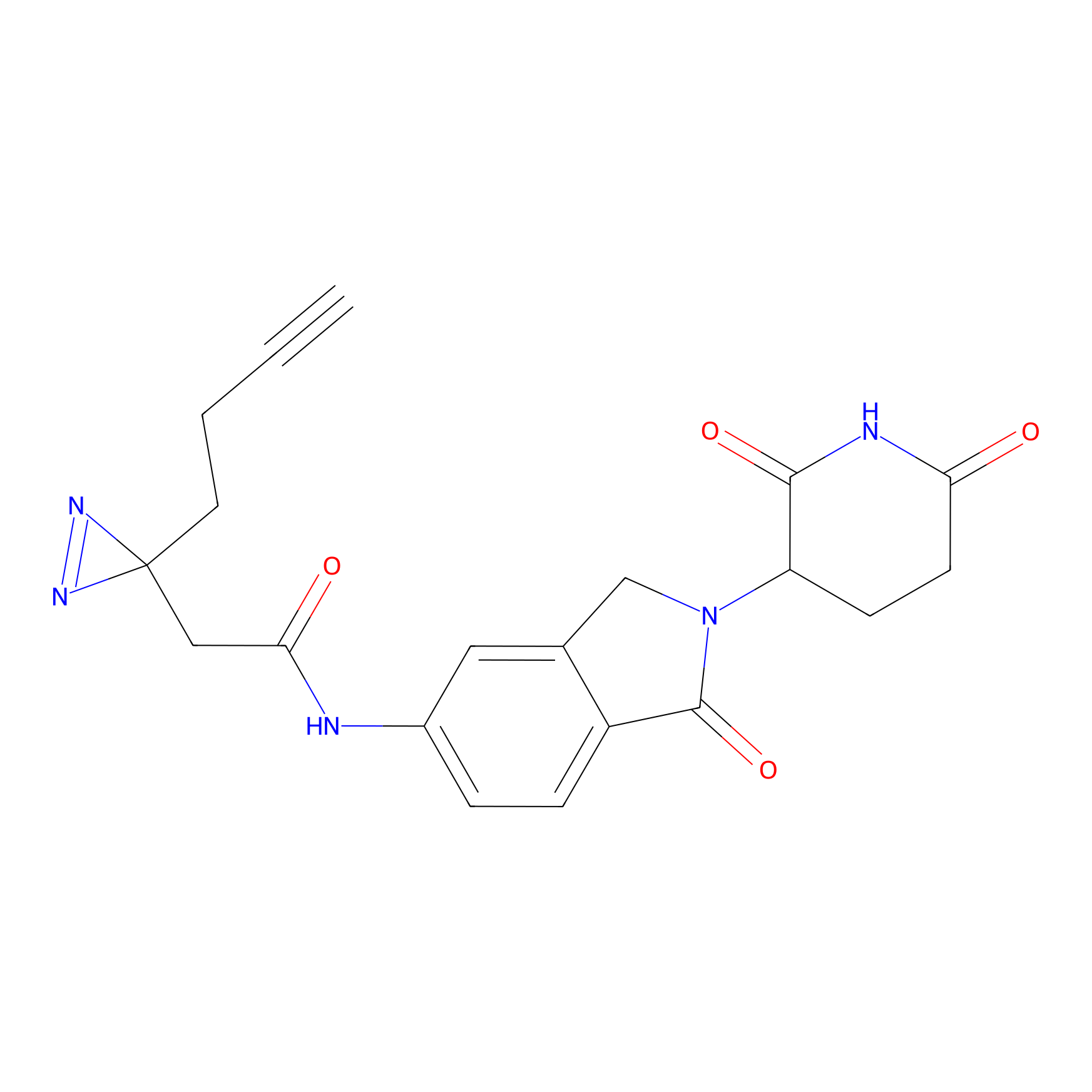 |
2.23 | LDD0256 | [10] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0116 | HHS-0101 | DM93 | Y106(0.82) | LDD0264 | [4] |
| LDCM0117 | HHS-0201 | DM93 | Y106(0.73) | LDD0265 | [4] |
| LDCM0118 | HHS-0301 | DM93 | Y106(0.71) | LDD0266 | [4] |
| LDCM0119 | HHS-0401 | DM93 | Y106(0.73) | LDD0267 | [4] |
| LDCM0120 | HHS-0701 | DM93 | Y106(0.60) | LDD0268 | [4] |
| LDCM0107 | IAA | HeLa | N.A. | LDD0221 | [6] |
| LDCM0115 | Minimalist | HEK-293T | 2.23 | LDD0256 | [10] |
| LDCM0109 | NEM | HeLa | N.A. | LDD0223 | [6] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Eukaryotic translation initiation factor 5B (EIF5B) | . | O60841 | |||
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Huntingtin (HTT) | Huntingtin family | P42858 | |||
Other
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Ataxin-1 (ATXN1) | ATXN1 family | P54253 | |||
References
