Details of the Target
General Information of Target
Target Site Mutations in Different Cell Lines
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
TH214 Probe Info |
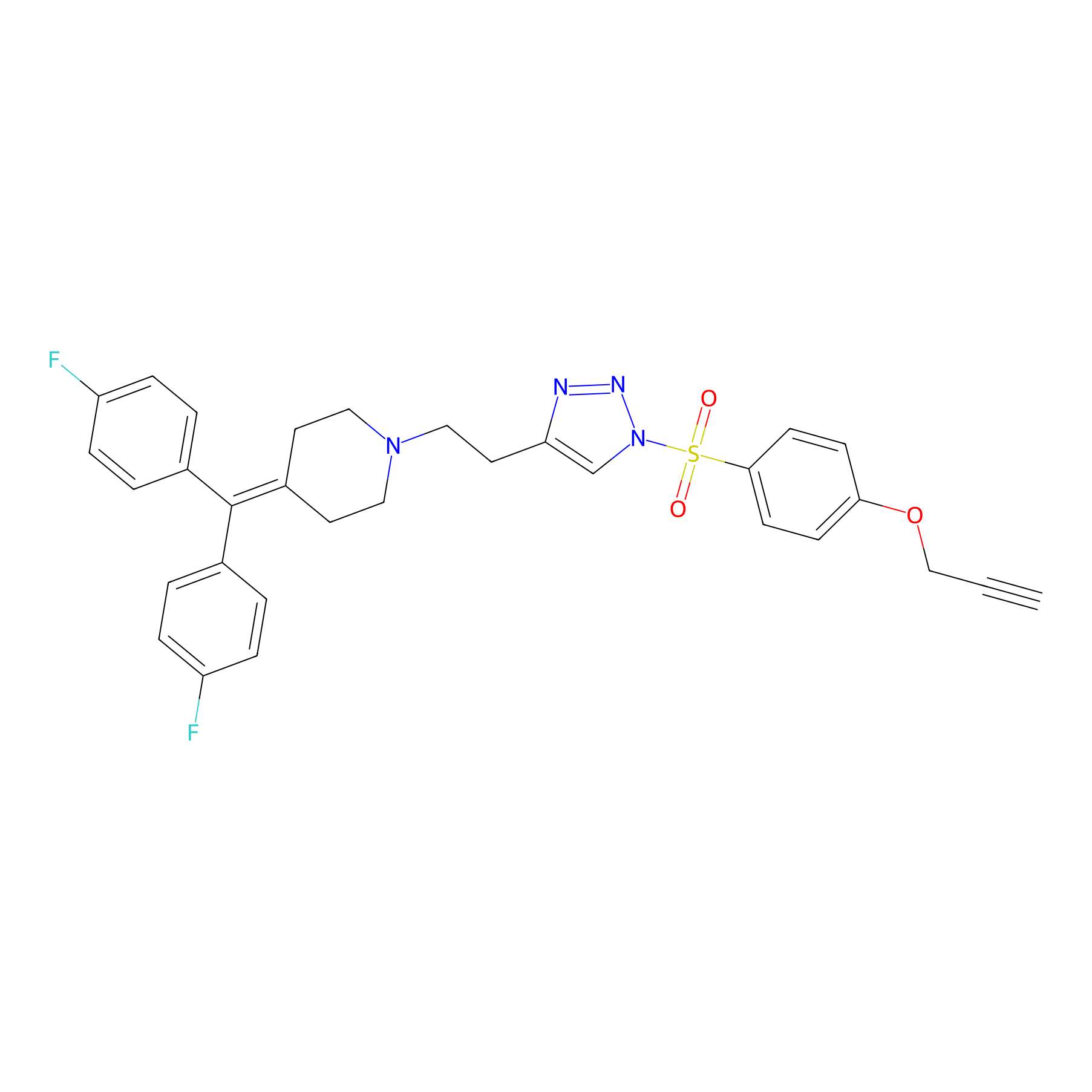 |
Y71(20.00) | LDD0258 | [1] | |
|
STPyne Probe Info |
 |
K14(3.39); K27(7.46) | LDD0277 | [2] | |
|
Probe 1 Probe Info |
 |
Y71(13.98) | LDD3495 | [3] | |
|
DBIA Probe Info |
 |
C70(1.81) | LDD3377 | [4] | |
|
BTD Probe Info |
 |
C45(1.06) | LDD2113 | [5] | |
|
HHS-475 Probe Info |
 |
Y71(0.43) | LDD0264 | [6] | |
|
HHS-465 Probe Info |
 |
Y122(10.00) | LDD2237 | [7] | |
|
ATP probe Probe Info |
 |
K75(0.00); K119(0.00); K47(0.00); K50(0.00) | LDD0199 | [8] | |
|
m-APA Probe Info |
 |
N.A. | LDD2234 | [9] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0162 | [10] | |
|
1d-yne Probe Info |
 |
N.A. | LDD0356 | [11] | |
|
NHS Probe Info |
 |
K75(0.00); K14(0.00) | LDD0010 | [12] | |
|
SF Probe Info |
 |
K32(0.00); K26(0.00); K14(0.00) | LDD0028 | [13] | |
|
1c-yne Probe Info |
 |
N.A. | LDD0228 | [11] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0217 | [14] | |
|
Methacrolein Probe Info |
 |
N.A. | LDD0218 | [14] | |
|
NAIA_5 Probe Info |
 |
N.A. | LDD2223 | [15] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C326 Probe Info |
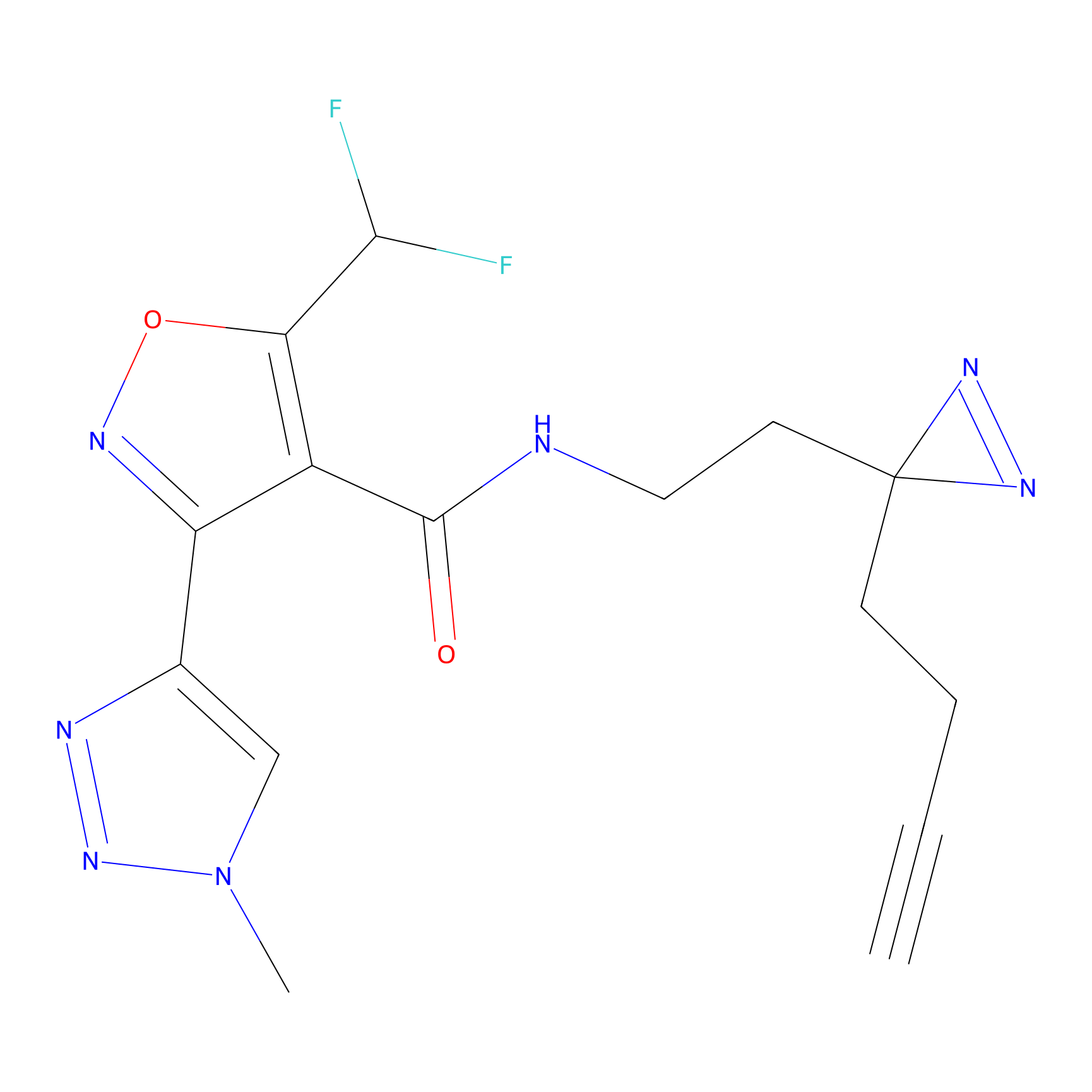 |
7.57 | LDD1990 | [16] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0520 | AKOS000195272 | MDA-MB-231 | C45(1.06) | LDD2113 | [5] |
| LDCM0116 | HHS-0101 | DM93 | Y71(0.43) | LDD0264 | [6] |
| LDCM0117 | HHS-0201 | DM93 | Y71(0.61) | LDD0265 | [6] |
| LDCM0118 | HHS-0301 | DM93 | Y71(0.30) | LDD0266 | [6] |
| LDCM0119 | HHS-0401 | DM93 | Y71(0.59) | LDD0267 | [6] |
| LDCM0120 | HHS-0701 | DM93 | Y71(0.83) | LDD0268 | [6] |
| LDCM0022 | KB02 | ICC108 | C70(1.33) | LDD2381 | [4] |
| LDCM0023 | KB03 | ICC108 | C70(1.53) | LDD2798 | [4] |
| LDCM0024 | KB05 | OPM-2 | C70(1.81) | LDD3377 | [4] |
| LDCM0109 | NEM | HeLa | N.A. | LDD0224 | [14] |
| LDCM0527 | Nucleophilic fragment 26b | MDA-MB-231 | C45(1.01) | LDD2120 | [5] |
| LDCM0535 | Nucleophilic fragment 30b | MDA-MB-231 | C45(0.98) | LDD2128 | [5] |
| LDCM0549 | Nucleophilic fragment 43 | MDA-MB-231 | C45(1.43) | LDD2143 | [5] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Transcription factor
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Zinc finger and BTB domain-containing protein 9 (ZBTB9) | . | Q96C00 | |||
Other
References
