Details of the Target
General Information of Target
| Target ID | LDTP12647 | |||||
|---|---|---|---|---|---|---|
| Target Name | Serine palmitoyltransferase 3 (SPTLC3) | |||||
| Gene Name | SPTLC3 | |||||
| Gene ID | 55304 | |||||
| Synonyms |
C20orf38; SPTLC2L; Serine palmitoyltransferase 3; EC 2.3.1.50; Long chain base biosynthesis protein 2b; LCB2b; Long chain base biosynthesis protein 3; LCB 3; Serine-palmitoyl-CoA transferase 3; SPT 3 |
|||||
| 3D Structure | ||||||
| Sequence |
MATDSWALAVDEQEAAVKSMTNLQIKEEKVKADTNGIIKTSTTAEKTDEEEKEDRAAQSL
LNKLIRSNLVDNTNQVEVLQRDPNSPLYSVKSFEELRLKPQLLQGVYAMGFNRPSKIQEN ALPMMLAEPPQNLIAQSQSGTGKTAAFVLAMLSRVEPSDRYPQCLCLSPTYELALQTGKV IEQMGKFYPELKLAYAVRGNKLERGQKISEQIVIGTPGTVLDWCSKLKFIDPKKIKVFVL DEADVMIATQGHQDQSIRIQRMLPRNCQMLLFSATFEDSVWKFAQKVVPDPNVIKLKREE ETLDTIKQYYVLCSSRDEKFQALCNLYGAITIAQAMIFCHTRKTASWLAAELSKEGHQVA LLSGEMMVEQRAAVIERFREGKEKVLVTTNVCARGIDVEQVSVVINFDLPVDKDGNPDNE TYLHRIGRTGRFGKRGLAVNMVDSKHSMNILNRIQEHFNKKIERLDTDDLDEIEKIAN |
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
Class-II pyridoxal-phosphate-dependent aminotransferase family
|
|||||
| Subcellular location |
Endoplasmic reticulum membrane
|
|||||
| Function |
Component of the serine palmitoyltransferase multisubunit enzyme (SPT) that catalyzes the initial and rate-limiting step in sphingolipid biosynthesis by condensing L-serine and activated acyl-CoA (most commonly palmitoyl-CoA) to form long-chain bases. The SPT complex is composed of SPTLC1, SPTLC2 or SPTLC3 and SPTSSA or SPTSSB. Within this complex, the heterodimer consisting of SPTLC1 and SPTLC2/SPTLC3 forms the catalytic core. The composition of the serine palmitoyltransferase (SPT) complex determines the substrate preference. The SPTLC1-SPTLC2-SPTSSA complex shows a strong preference for C16-CoA substrate, while the SPTLC1-SPTLC3-SPTSSA isozyme uses both C14-CoA and C16-CoA as substrates, with a slight preference for C14-CoA. The SPTLC1-SPTLC2-SPTSSB complex shows a strong preference for C18-CoA substrate, while the SPTLC1-SPTLC3-SPTSSB isozyme displays an ability to use a broader range of acyl-CoAs, without apparent preference.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
| Cell line | Mutation details | Probe for labeling this protein in this cell | |||
|---|---|---|---|---|---|
| CCK81 | SNV: p.A343G | DBIA Probe Info | |||
| IPC298 | SNV: p.T493P | . | |||
| MCC13 | SNV: p.L495F | DBIA Probe Info | |||
| NCIH2172 | SNV: p.I412M | . | |||
| OVCAR5 | SNV: p.A472V | . | |||
| OVK18 | SNV: p.I483N | . | |||
| SNU869 | SNV: p.F355S | . | |||
| SW480 | SNV: p.N254I | . | |||
| SW620 | SNV: p.N254I | . | |||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
IPM Probe Info |
 |
C91(0.00); C10(0.00) | LDD0241 | [1] | |
|
DBIA Probe Info |
 |
C91(2.10) | LDD3332 | [2] | |
|
BTD Probe Info |
 |
C503(1.13) | LDD2098 | [3] | |
|
Methacrolein Probe Info |
 |
N.A. | LDD0218 | [4] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
Alk-rapa Probe Info |
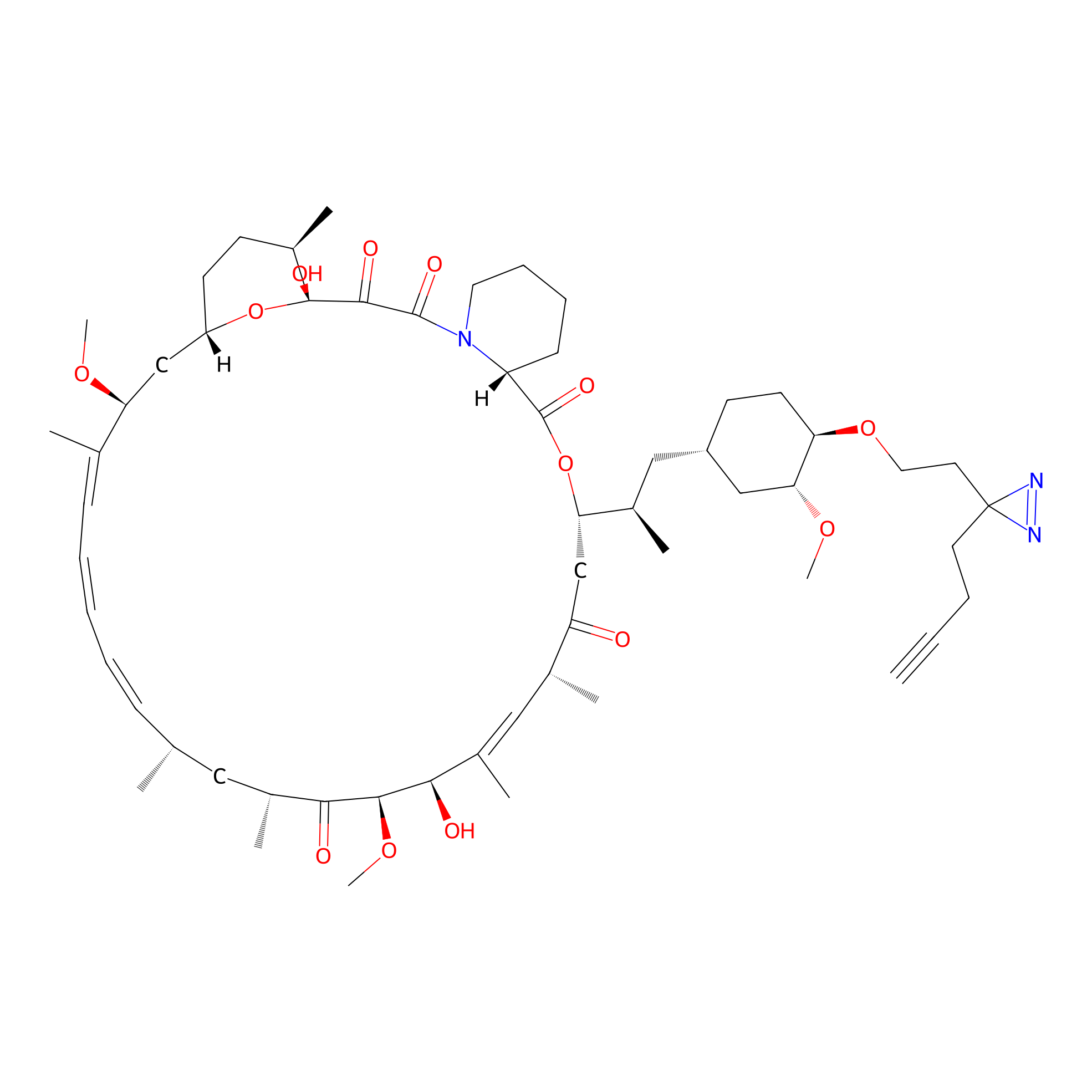 |
3.28 | LDD0213 | [5] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0022 | KB02 | 42-MG-BA | C91(2.57) | LDD2244 | [2] |
| LDCM0023 | KB03 | 42-MG-BA | C91(1.84) | LDD2661 | [2] |
| LDCM0024 | KB05 | MKN45 | C91(2.10) | LDD3332 | [2] |
| LDCM0505 | Nucleophilic fragment 15b | MDA-MB-231 | C503(1.13) | LDD2098 | [3] |
| LDCM0512 | Nucleophilic fragment 19a | MDA-MB-231 | C503(1.02) | LDD2105 | [3] |
| LDCM0090 | Rapamycin | JHH-7 | 3.28 | LDD0213 | [5] |
The Interaction Atlas With This Target
References
