Details of the Target
General Information of Target
| Target ID | LDTP12267 | |||||
|---|---|---|---|---|---|---|
| Target Name | Cytochrome P450 4F12 (CYP4F12) | |||||
| Gene Name | CYP4F12 | |||||
| Gene ID | 66002 | |||||
| Synonyms |
Cytochrome P450 4F12; EC 1.14.14.1; CYPIVF12 |
|||||
| 3D Structure | ||||||
| Sequence |
MAASRLDFGEVETFLDRHPELFEDYLMRKGKQEMVEKWLQRHSQGQGALGPRPSLAGTSS
LAHSTCRGGSSVGGGTGPNGSAHSQPLPGGGDCGGVPLSPSWAGGSRGDGNLQRRASQKE LRKSFARSKAIHVNRTYDEQVTSRAQEPLSSVRRRALLRKASSLPPTTAHILSALLESRV NLPRYPPTAIDYKCHLKKHNERQFFLELVKDISNDLDLTSLSYKILIFVCLMVDADRCSL FLVEGAAAGKKTLVSKFFDVHAGTPLLPCSSTENSNEVQVPWGKGIIGYVGEHGETVNIP DAYQDRRFNDEIDKLTGYKTKSLLCMPIRSSDGEIIGVAQAINKIPEGAPFTEDDEKVMQ MYLPFCGIAISNAQLFAASRKEYERSRALLEVVNDLFEEQTDLEKIVKKIMHRAQTLLKC ERCSVLLLEDIESPVVKFTKSFELMSPKCSADAENSFKESMEKSSYSDWLINNSIAELVA STGLPVNISDAYQDPRFDAEADQISGFHIRSVLCVPIWNSNHQIIGVAQVLNRLDGKPFD DADQRLFEAFVIFCGLGINNTIMYDQVKKSWAKQSVALDVLSYHATCSKAEVDKFKAANI PLVSELAIDDIHFDDFSLDVDAMITAALRMFMELGMVQKFKIDYETLCRWLLTVRKNYRM VLYHNWRHAFNVCQLMFAMLTTAGFQDILTEVEILAVIVGCLCHDLDHRGTNNAFQAKSG SALAQLYGTSATLEHHHFNHAVMILQSEGHNIFANLSSKEYSDLMQLLKQSILATDLTLY FERRTEFFELVSKGEYDWNIKNHRDIFRSMLMTACDLGAVTKPWEISRQVAELVTSEFFE QGDRERLELKLTPSAIFDRNRKDELPRLQLEWIDSICMPLYQALVKVNVKLKPMLDSVAT NRSKWEELHQKRLLASTASSSPASVMVAKEDRN |
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
Cytochrome P450 family
|
|||||
| Subcellular location |
Endoplasmic reticulum membrane
|
|||||
| Function |
A cytochrome P450 monooxygenase involved in the metabolism of endogenous polyunsaturated fatty acids (PUFAs). Mechanistically, uses molecular oxygen inserting one oxygen atom into a substrate, and reducing the second into a water molecule, with two electrons provided by NADPH via cytochrome P450 reductase (CPR; NADPH-ferrihemoprotein reductase). Catalyzes the hydroxylation of carbon hydrogen bonds, with preference for omega-2 position. Metabolizes (5Z,8Z,11Z,14Z)-eicosatetraenoic acid (arachidonate) toward 18-hydroxy arachidonate. Catalyzes the epoxidation of double bonds of PUFAs such as docosapentaenoic and docosahexaenoic acids. Has low omega-hydroxylase activity toward leukotriene B4 and arachidonate. Involved in the metabolism of xenobiotics. Catalyzes the hydroxylation of the antihistamine drug ebastine.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
| ChEMBL ID | ||||||
Target Site Mutations in Different Cell Lines
| Cell line | Mutation details | Probe for labeling this protein in this cell | |||
|---|---|---|---|---|---|
| CORL23 | SNV: p.L228F | . | |||
| HT | SNV: p.S314P | . | |||
| KMCH1 | SNV: p.R365L | . | |||
| MELHO | Deletion: p.D142_K143del SNV: p.W144G |
. | |||
| MEWO | SNV: p.P396S | . | |||
| MFE319 | SNV: p.G328R | . | |||
| NCIH2170 | SNV: p.M80I | DBIA Probe Info | |||
| REH | SNV: p.A182T | . | |||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
IPM Probe Info |
 |
C50(0.00); C401(0.00); C402(0.00) | LDD0241 | [1] | |
|
DBIA Probe Info |
 |
C50(2.53); C468(1.25) | LDD3321 | [2] | |
|
WYneN Probe Info |
 |
N.A. | LDD0021 | [3] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
AEA-DA Probe Info |
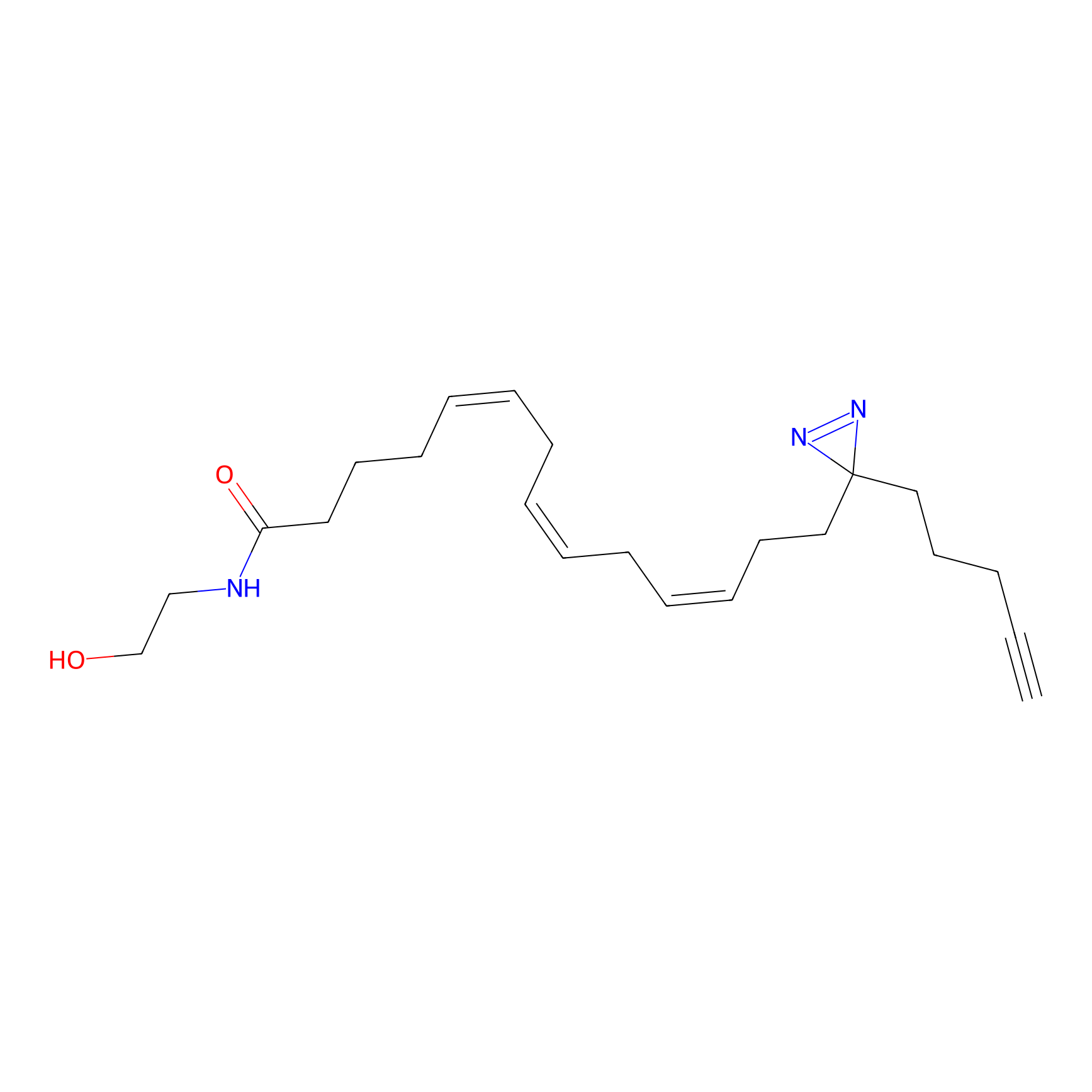 |
2.42 | LDD0146 | [4] | |
Competitor(s) Related to This Target
The Interaction Atlas With This Target
References
