Details of the Target
General Information of Target
| Target ID | LDTP09705 | |||||
|---|---|---|---|---|---|---|
| Target Name | Connector enhancer of kinase suppressor of ras 2 (CNKSR2) | |||||
| Gene Name | CNKSR2 | |||||
| Gene ID | 22866 | |||||
| Synonyms |
CNK2; KIAA0902; KSR2; Connector enhancer of kinase suppressor of ras 2; Connector enhancer of KSR 2; CNK homolog protein 2; CNK2 |
|||||
| 3D Structure | ||||||
| Sequence |
MALIMEPVSKWSPSQVVDWMKGLDDCLQQYIKNFEREKISGDQLLRITHQELEDLGVSRI
GHQELILEAVDLLCALNYGLETENLKTLSHKLNASAKNLQNFITGRRRSGHYDGRTSRKL PNDFLTSVVDLIGAAKSLLAWLDRSPFAAVTDYSVTRNNVIQLCLELTTIVQQDCTVYET ENKILHVCKTLSGVCDHIISLSSDPLVSQSAHLEVIQLANIKPSEGLGMYIKSTYDGLHV ITGTTENSPADRCKKIHAGDEVIQVNHQTVVGWQLKNLVNALREDPSGVILTLKKRPQSM LTSAPALLKNMRWKPLALQPLIPRSPTSSVATPSSTISTPTKRDSSALQDLYIPPPPAEP YIPRDEKGNLPCEDLRGHMVGKPVHKGSESPNSFLDQEYRKRFNIVEEDTVLYCYEYEKG RSSSQGRRESTPTYGKLRPISMPVEYNWVGDYEDPNKMKRDSRRENSLLRYMSNEKIAQE EYMFQRNSKKDTGKKSKKKGDKSNSPTHYSLLPSLQMDALRQDIMGTPVPETTLYHTFQQ SSLQHKSKKKNKGPIAGKSKRRISCKDLGRGDCEGWLWKKKDAKSYFSQKWKKYWFVLKD ASLYWYINEEDEKAEGFISLPEFKIDRASECRKKYAFKACHPKIKSFYFAAEHLDDMNRW LNRINMLTAGYAERERIKQEQDYWSESDKEEADTPSTPKQDSPPPPYDTYPRPPSMSCAS PYVEAKHSRLSSTETSQSQSSHEEFRQEVTGSSAVSPIRKTASQRRSWQDLIETPLTSSG LHYLQTLPLEDSVFSDSAAISPEHRRQSTLPTQKCHLQDHYGPYPLAESERMQVLNGNGG KPRSFTLPRDSGFNHCCLNAPVSACDPQDDVQPPEVEEEEEEEEEEGEAAGENIGEKSES REEKLGDSLQDLYRALEQASLSPLGEHRISTKMEYKLSFIKRCNDPVMNEKLHRLRILKS TLKAREGEVAIIDKVLDNPDLTSKEFQQWKQMYLDLFLDICQNTTSNDPLSISSEVDVIT SSLAHTHSYIETHV |
|||||
| Target Bioclass |
Other
|
|||||
| Family |
CNKSR family
|
|||||
| Subcellular location |
Cytoplasm
|
|||||
| Function | May function as an adapter protein or regulator of Ras signaling pathways. | |||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
| ChEMBL ID | ||||||
Target Site Mutations in Different Cell Lines
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
P1 Probe Info |
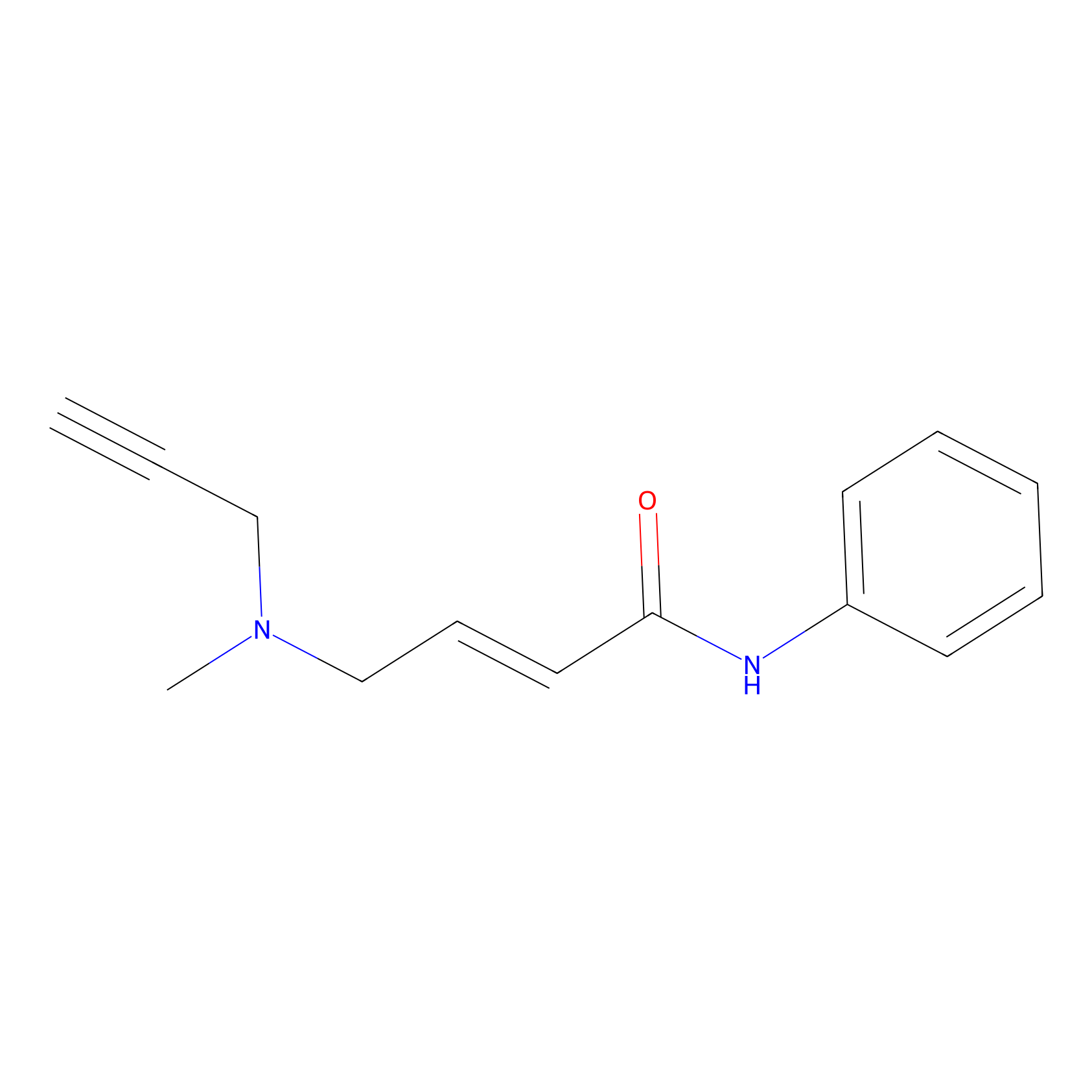 |
3.32 | LDD0448 | [1] | |
|
YN-1 Probe Info |
 |
100.00 | LDD0444 | [2] | |
|
DBIA Probe Info |
 |
C573(2.02) | LDD3410 | [3] | |
Competitor(s) Related to This Target
References
