Details of the Target
General Information of Target
| Target ID | LDTP06215 | |||||
|---|---|---|---|---|---|---|
| Target Name | Myocyte-specific enhancer factor 2D (MEF2D) | |||||
| Gene Name | MEF2D | |||||
| Gene ID | 4209 | |||||
| Synonyms |
Myocyte-specific enhancer factor 2D |
|||||
| 3D Structure | ||||||
| Sequence |
MGRKKIQIQRITDERNRQVTFTKRKFGLMKKAYELSVLCDCEIALIIFNHSNKLFQYAST
DMDKVLLKYTEYNEPHESRTNADIIETLRKKGFNGCDSPEPDGEDSLEQSPLLEDKYRRA SEELDGLFRRYGSTVPAPNFAMPVTVPVSNQSSLQFSNPSGSLVTPSLVTSSLTDPRLLS PQQPALQRNSVSPGLPQRPASAGAMLGGDLNSANGACPSPVGNGYVSARASPGLLPVANG NSLNKVIPAKSPPPPTHSTQLGAPSRKPDLRVITSQAGKGLMHHLTEDHLDLNNAQRLGV SQSTHSLTTPVVSVATPSLLSQGLPFSSMPTAYNTDYQLTSAELSSLPAFSSPGGLSLGN VTAWQQPQQPQQPQQPQPPQQQPPQPQQPQPQQPQQPQQPPQQQSHLVPVSLSNLIPGSP LPHVGAALTVTTHPHISIKSEPVSPSRERSPAPPPPAVFPAARPEPGDGLSSPAGGSYET GDRDDGRGDFGPTLGLLRPAPEPEAEGSAVKRMRLDTWTLK |
|||||
| Target Bioclass |
Transcription factor
|
|||||
| Family |
MEF2 family
|
|||||
| Subcellular location |
Nucleus
|
|||||
| Function |
Transcriptional activator which binds specifically to the MEF2 element, 5'-YTA[AT](4)TAR-3', found in numerous muscle-specific, growth factor- and stress-induced genes. Mediates cellular functions not only in skeletal and cardiac muscle development, but also in neuronal differentiation and survival. Plays diverse roles in the control of cell growth, survival and apoptosis via p38 MAPK signaling in muscle-specific and/or growth factor-related transcription. Plays a critical role in the regulation of neuronal apoptosis.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
15.00 | LDD0402 | [1] | |
|
EA-probe Probe Info |
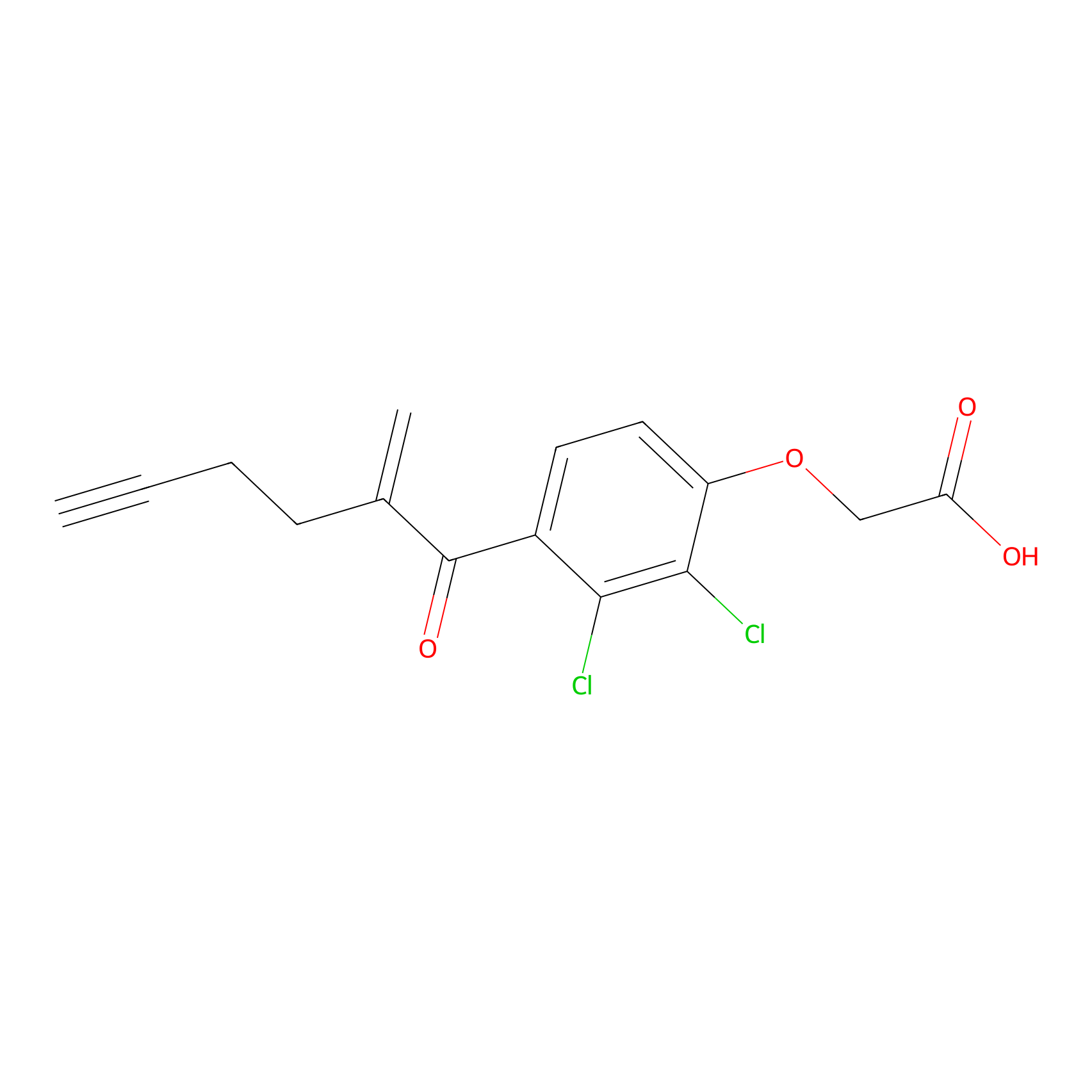 |
N.A. | LDD0440 | [2] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0227 | [3] | |
|
NAIA_4 Probe Info |
 |
N.A. | LDD2226 | [4] | |
|
AOyne Probe Info |
 |
9.80 | LDD0443 | [5] | |
Competitor(s) Related to This Target
References
