Details of the Target
General Information of Target
| Target ID | LDTP04941 | |||||
|---|---|---|---|---|---|---|
| Target Name | Coatomer subunit zeta-1 (COPZ1) | |||||
| Gene Name | COPZ1 | |||||
| Gene ID | 22818 | |||||
| Synonyms |
COPZ; Coatomer subunit zeta-1; Zeta-1-coat protein; Zeta-1 COP |
|||||
| 3D Structure | ||||||
| Sequence |
MEALILEPSLYTVKAILILDNDGDRLFAKYYDDTYPSVKEQKAFEKNIFNKTHRTDSEIA
LLEGLTVVYKSSIDLYFYVIGSSYENELMLMAVLNCLFDSLSQMLRKNVEKRALLENMEG LFLAVDEIVDGGVILESDPQQVVHRVALRGEDVPLTEQTVSQVLQSAKEQIKWSLLR |
|||||
| Target Bioclass |
Other
|
|||||
| Family |
Adaptor complexes small subunit family
|
|||||
| Subcellular location |
Cytoplasm
|
|||||
| Function |
The coatomer is a cytosolic protein complex that binds to dilysine motifs and reversibly associates with Golgi non-clathrin-coated vesicles, which further mediate biosynthetic protein transport from the ER, via the Golgi up to the trans Golgi network. Coatomer complex is required for budding from Golgi membranes, and is essential for the retrograde Golgi-to-ER transport of dilysine-tagged proteins. The zeta subunit may be involved in regulating the coat assembly and, hence, the rate of biosynthetic protein transport due to its association-dissociation properties with the coatomer complex.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
9.27 | LDD0402 | [1] | |
|
TH216 Probe Info |
 |
Y35(11.48) | LDD0259 | [2] | |
|
ONAyne Probe Info |
 |
K51(5.41) | LDD0274 | [3] | |
|
Probe 1 Probe Info |
 |
Y31(126.77); Y35(22.08) | LDD3495 | [4] | |
|
ATP probe Probe Info |
 |
K39(0.00); K42(0.00) | LDD0199 | [5] | |
|
STPyne Probe Info |
 |
N.A. | LDD0009 | [6] | |
|
1c-yne Probe Info |
 |
N.A. | LDD0228 | [7] | |
|
AOyne Probe Info |
 |
6.10 | LDD0443 | [8] | |
|
TER-AC Probe Info |
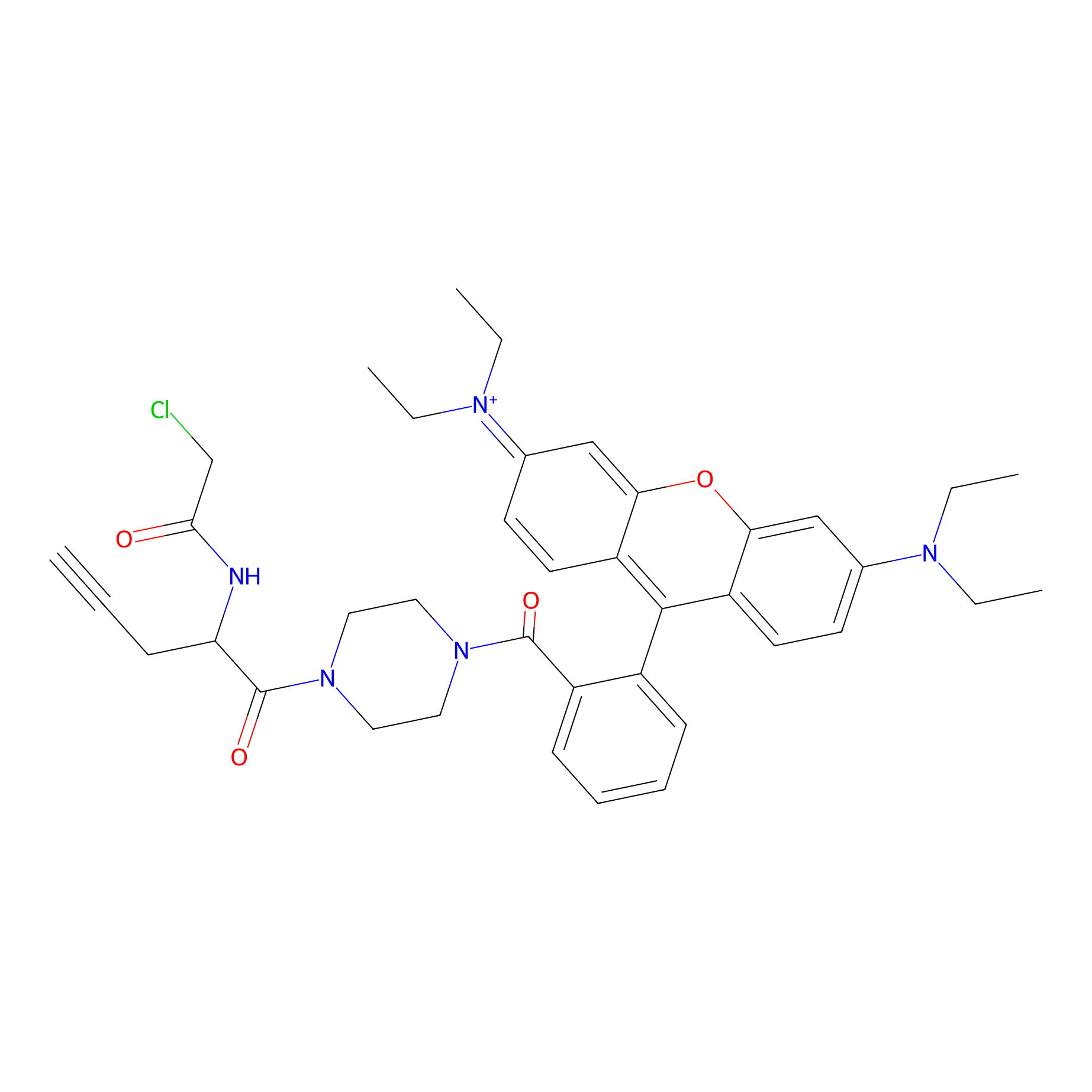 |
N.A. | LDD0426 | [9] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C040 Probe Info |
 |
7.94 | LDD1740 | [10] | |
Competitor(s) Related to This Target
References
