Details of the Target
General Information of Target
| Target ID | LDTP02895 | |||||
|---|---|---|---|---|---|---|
| Target Name | Atrial natriuretic peptide receptor 3 (NPR3) | |||||
| Gene Name | NPR3 | |||||
| Gene ID | 4883 | |||||
| Synonyms |
ANPRC; C5orf23; NPRC; Atrial natriuretic peptide receptor 3; Atrial natriuretic peptide clearance receptor; Atrial natriuretic peptide receptor type C; ANP-C; ANPR-C; NPR-C |
|||||
| 3D Structure | ||||||
| Sequence |
MPSLLVLTFSPCVLLGWALLAGGTGGGGVGGGGGGAGIGGGRQEREALPPQKIEVLVLLP
QDDSYLFSLTRVRPAIEYALRSVEGNGTGRRLLPPGTRFQVAYEDSDCGNRALFSLVDRV AAARGAKPDLILGPVCEYAAAPVARLASHWDLPMLSAGALAAGFQHKDSEYSHLTRVAPA YAKMGEMMLALFRHHHWSRAALVYSDDKLERNCYFTLEGVHEVFQEEGLHTSIYSFDETK DLDLEDIVRNIQASERVVIMCASSDTIRSIMLVAHRHGMTSGDYAFFNIELFNSSSYGDG SWKRGDKHDFEAKQAYSSLQTVTLLRTVKPEFEKFSMEVKSSVEKQGLNMEDYVNMFVEG FHDAILLYVLALHEVLRAGYSKKDGGKIIQQTWNRTFEGIAGQVSIDANGDRYGDFSVIA MTDVEAGTQEVIGDYFGKEGRFEMRPNVKYPWGPLKLRIDENRIVEHTNSSPCKSSGGLE ESAVTGIVVGALLGAGLLMAFYFFRKKYRITIERRTQQEESNLGKHRELREDSIRSHFSV A |
|||||
| Target Type |
Clinical trial
|
|||||
| Target Bioclass |
Other
|
|||||
| Family |
ANF receptor family
|
|||||
| Subcellular location |
Cell membrane
|
|||||
| Function |
Receptor for the natriuretic peptide hormones, binding with similar affinities atrial natriuretic peptide NPPA/ANP, brain natriuretic peptide NPPB/BNP, and C-type natriuretic peptide NPPC/CNP. May function as a clearance receptor for NPPA, NPPB and NPPC, regulating their local concentrations and effects. May regulate diuresis, blood pressure and skeletal development. Does not have guanylate cyclase activity.
|
|||||
| TTD ID | ||||||
| Uniprot ID | ||||||
| DrugMap ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
| ChEMBL ID | ||||||
Target Site Mutations in Different Cell Lines
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
FBPP2 Probe Info |
 |
4.07 | LDD0324 | [1] | |
|
OPA-S-S-alkyne Probe Info |
 |
K456(0.78); K127(4.43); K208(5.04) | LDD3494 | [2] | |
|
DBIA Probe Info |
 |
C136(2.73); C473(4.71) | LDD3365 | [3] | |
|
IPM Probe Info |
 |
N.A. | LDD0147 | [4] | |
|
TFBX Probe Info |
 |
N.A. | LDD0148 | [4] | |
|
Acrolein Probe Info |
 |
C473(0.00); C108(0.00); H467(0.00) | LDD0217 | [5] | |
|
Cinnamaldehyde Probe Info |
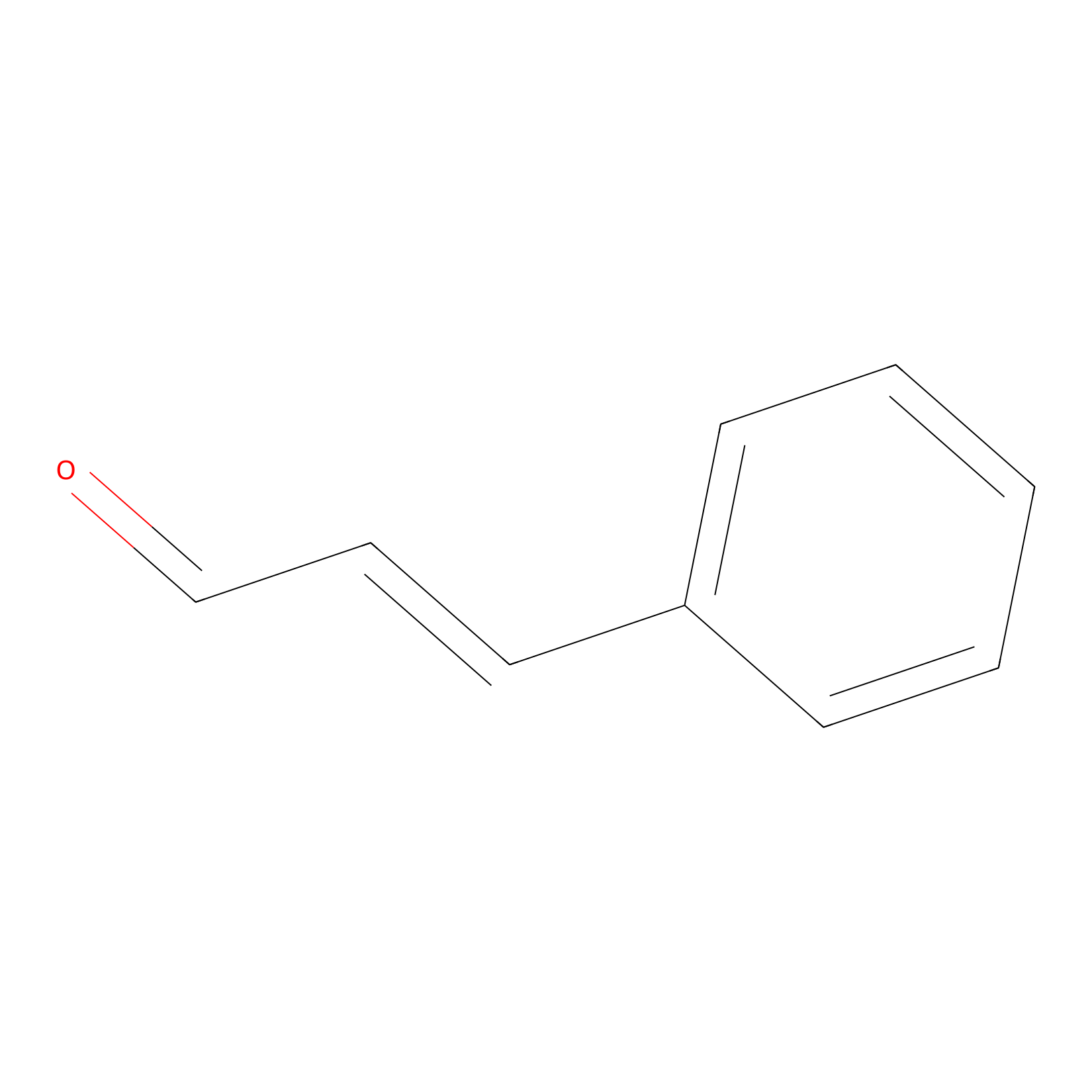 |
N.A. | LDD0220 | [5] | |
|
Crotonaldehyde Probe Info |
 |
H173(0.00); C108(0.00) | LDD0219 | [5] | |
|
Methacrolein Probe Info |
 |
C473(0.00); C108(0.00); C261(0.00) | LDD0218 | [5] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0108 | Chloroacetamide | HeLa | C108(0.00); C473(0.00) | LDD0222 | [5] |
| LDCM0107 | IAA | HeLa | N.A. | LDD0221 | [5] |
| LDCM0022 | KB02 | EFO-27 | C136(1.25) | LDD2322 | [3] |
| LDCM0023 | KB03 | ICC108 | C136(2.02) | LDD2798 | [3] |
| LDCM0024 | KB05 | OVCAR-3 | C136(2.73); C473(4.71) | LDD3365 | [3] |
| LDCM0109 | NEM | HeLa | N.A. | LDD0223 | [5] |
The Interaction Atlas With This Target
References
