Details of the Target
General Information of Target
| Target ID | LDTP02812 | |||||
|---|---|---|---|---|---|---|
| Target Name | Beta-galactoside alpha-2,6-sialyltransferase 1 (ST6GAL1) | |||||
| Gene Name | ST6GAL1 | |||||
| Gene ID | 6480 | |||||
| Synonyms |
SIAT1; Beta-galactoside alpha-2,6-sialyltransferase 1; Alpha 2,6-ST 1; EC 2.4.3.1; B-cell antigen CD75; CMP-N-acetylneuraminate-beta-galactosamide-alpha-2,6-sialyltransferase 1; ST6Gal I; ST6GalI; Sialyltransferase 1
|
|||||
| 3D Structure | ||||||
| Sequence |
MIHTNLKKKFSCCVLVFLLFAVICVWKEKKKGSYYDSFKLQTKEFQVLKSLGKLAMGSDS
QSVSSSSTQDPHRGRQTLGSLRGLAKAKPEASFQVWNKDSSSKNLIPRLQKIWKNYLSMN KYKVSYKGPGPGIKFSAEALRCHLRDHVNVSMVEVTDFPFNTSEWEGYLPKESIRTKAGP WGRCAVVSSAGSLKSSQLGREIDDHDAVLRFNGAPTANFQQDVGTKTTIRLMNSQLVTTE KRFLKDSLYNEGILIVWDPSVYHSDIPKWYQNPDYNFFNNYKTYRKLHPNQPFYILKPQM PWELWDILQEISPEEIQPNPPSSGMLGIIIMMTLCDQVDIYEFLPSKRKTDVCYYYQKFF DSACTMGAYHPLLYEKNLVKHLNQGTDEDIYLLGKATLPGFRTIHC |
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
Glycosyltransferase 29 family
|
|||||
| Subcellular location |
Golgi apparatus, Golgi stack membrane
|
|||||
| Function | Transfers sialic acid from CMP-sialic acid to galactose-containing acceptor substrates. | |||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
| ChEMBL ID | ||||||
Target Site Mutations in Different Cell Lines
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
DBIA Probe Info |
 |
C184(1.82) | LDD3424 | [1] | |
|
Alkyne-RA190 Probe Info |
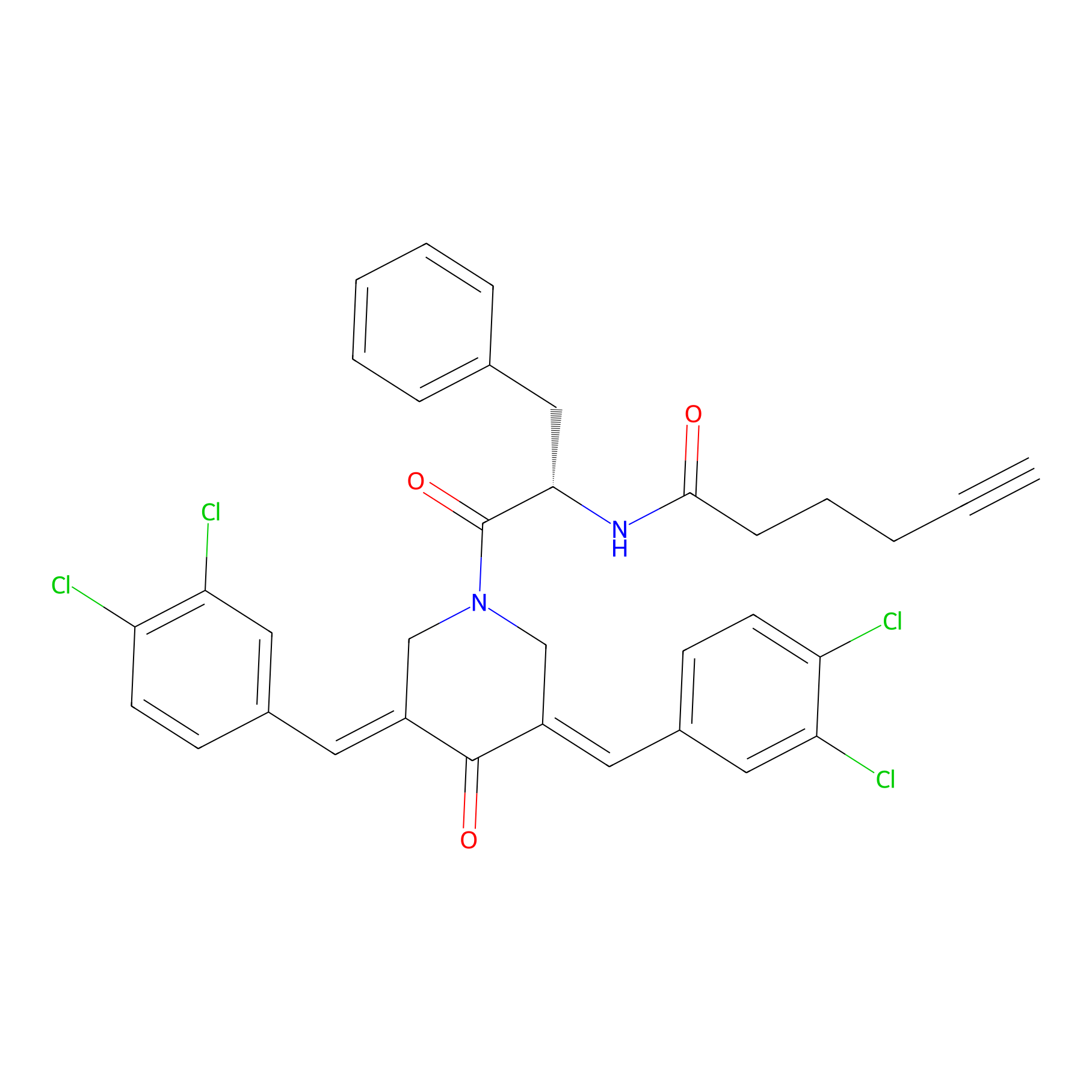 |
8.96 | LDD0299 | [2] | |
|
IA-alkyne Probe Info |
 |
C364(1.26) | LDD2185 | [3] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0625 | F8 | Ramos | C364(1.55) | LDD2187 | [3] |
| LDCM0573 | Fragment11 | Ramos | C364(0.15) | LDD2190 | [3] |
| LDCM0575 | Fragment13 | Ramos | C364(1.23) | LDD2192 | [3] |
| LDCM0576 | Fragment14 | Ramos | C364(1.74) | LDD2193 | [3] |
| LDCM0580 | Fragment21 | Ramos | C364(1.33) | LDD2195 | [3] |
| LDCM0582 | Fragment23 | Ramos | C364(1.11) | LDD2196 | [3] |
| LDCM0578 | Fragment27 | Ramos | C364(0.82) | LDD2197 | [3] |
| LDCM0586 | Fragment28 | Ramos | C364(1.75) | LDD2198 | [3] |
| LDCM0588 | Fragment30 | Ramos | C364(1.36) | LDD2199 | [3] |
| LDCM0468 | Fragment33 | Ramos | C364(1.03) | LDD2202 | [3] |
| LDCM0596 | Fragment38 | Ramos | C364(0.55) | LDD2203 | [3] |
| LDCM0610 | Fragment52 | Ramos | C364(1.36) | LDD2204 | [3] |
| LDCM0614 | Fragment56 | Ramos | C364(1.03) | LDD2205 | [3] |
| LDCM0569 | Fragment7 | Ramos | C364(2.01) | LDD2186 | [3] |
| LDCM0022 | KB02 | AU565 | C353(4.73) | LDD2265 | [1] |
| LDCM0023 | KB03 | AU565 | C353(9.00) | LDD2682 | [1] |
| LDCM0024 | KB05 | SK-HEP1 | C184(1.82) | LDD3424 | [1] |
| LDCM0131 | RA190 | MM1.R | 8.96 | LDD0299 | [2] |
References
