Details of the Target
General Information of Target
| Target ID | LDTP00114 | |||||
|---|---|---|---|---|---|---|
| Target Name | Putative annexin A2-like protein (ANXA2P2) | |||||
| Gene Name | ANXA2P2 | |||||
| Synonyms |
ANX2L2; ANX2P2; LPC2B; Putative annexin A2-like protein; Annexin A2 pseudogene 2; Lipocortin II pseudogene |
|||||
| 3D Structure | ||||||
| Sequence |
MSTVHEILCKLSLEGDHSTPPSAYGSVKAYTNFDAERDALNIETAIKTKGVDEVTIVNIV
TNRDNAQRQDIVFSYQRRTKKELASALKSALSGHLETVILGLLKTPAQYDASELKASMKG LGTDEDSLIEIICSRTNQELQEINRVYKEMYKTDLEKDIISDTSGDFRKLMVALAKGRRA EDGSVIDYELIDQDAQDLYDAGVKRKGTDVPKWISIMTERSVPHLQKVFDRYKSYSPYDM LESIRKEVKGDLENAFLNLVQRIQNKPLYFADQLYDSMKGKGTRDKVLIRIMVSRSEVDM LKIRSEFKRKYGKSLYYYIQQDTKGDYQKALLYLCGGDD |
|||||
| Target Bioclass |
Other
|
|||||
| Family |
Annexin family
|
|||||
| Subcellular location |
Secreted, extracellular space, extracellular matrix, basement membrane
|
|||||
| Function |
Calcium-regulated membrane-binding protein whose affinity for calcium is greatly enhanced by anionic phospholipids. It binds two calcium ions with high affinity. May be involved in heat-stress response.
|
|||||
| Uniprot ID | ||||||
| HGNC ID | ||||||
| ChEMBL ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
AZ-9 Probe Info |
 |
D322(0.97); D34(0.98); E36(0.98); E219(0.96) | LDD2208 | [1] | |
|
OPA-S-S-alkyne Probe Info |
 |
K212(1.42); K28(1.99); K119(2.12); K157(4.25) | LDD3494 | [2] | |
|
DBIA Probe Info |
 |
C353(4.39); C151(6.91) | LDD3312 | [3] | |
|
THZ1-DTB Probe Info |
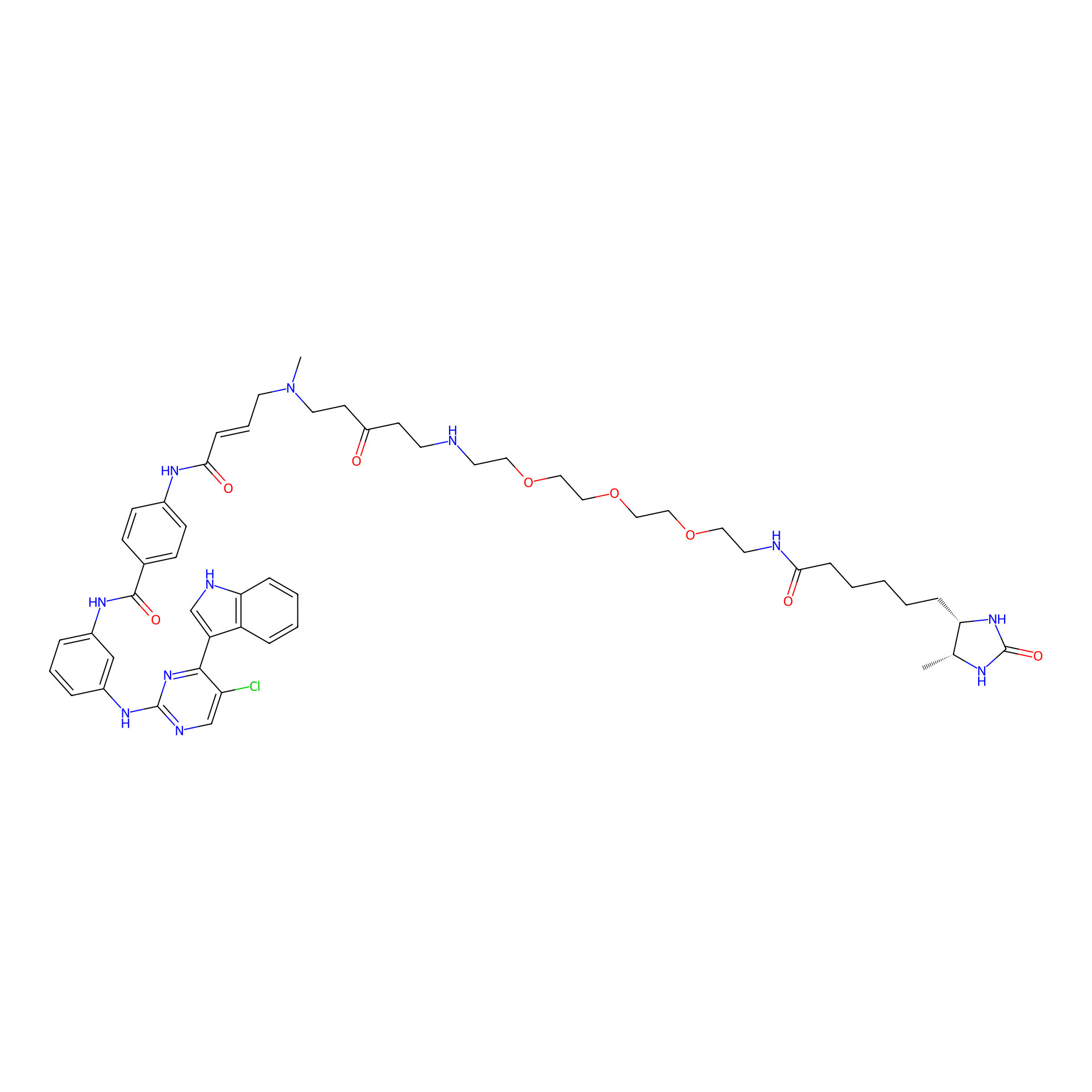 |
C133(1.10) | LDD0460 | [4] | |
|
HHS-475 Probe Info |
 |
Y316(0.74); Y317(0.81); Y24(0.86); Y235(1.01) | LDD0264 | [5] | |
|
IA-alkyne Probe Info |
 |
C9(0.59) | LDD2206 | [6] | |
|
HHS-465 Probe Info |
 |
Y238(10.00); Y24(1.56); Y30(10.00); Y317(5.56) | LDD2237 | [7] | |
|
5E-2FA Probe Info |
 |
H17(0.00); H94(0.00); H5(0.00) | LDD2235 | [8] | |
|
m-APA Probe Info |
 |
H17(0.00); H94(0.00); H5(0.00) | LDD2231 | [8] | |
|
JW-RF-010 Probe Info |
 |
C9(0.00); C133(0.00) | LDD0026 | [9] | |
|
IPM Probe Info |
 |
N.A. | LDD0005 | [10] | |
|
NPM Probe Info |
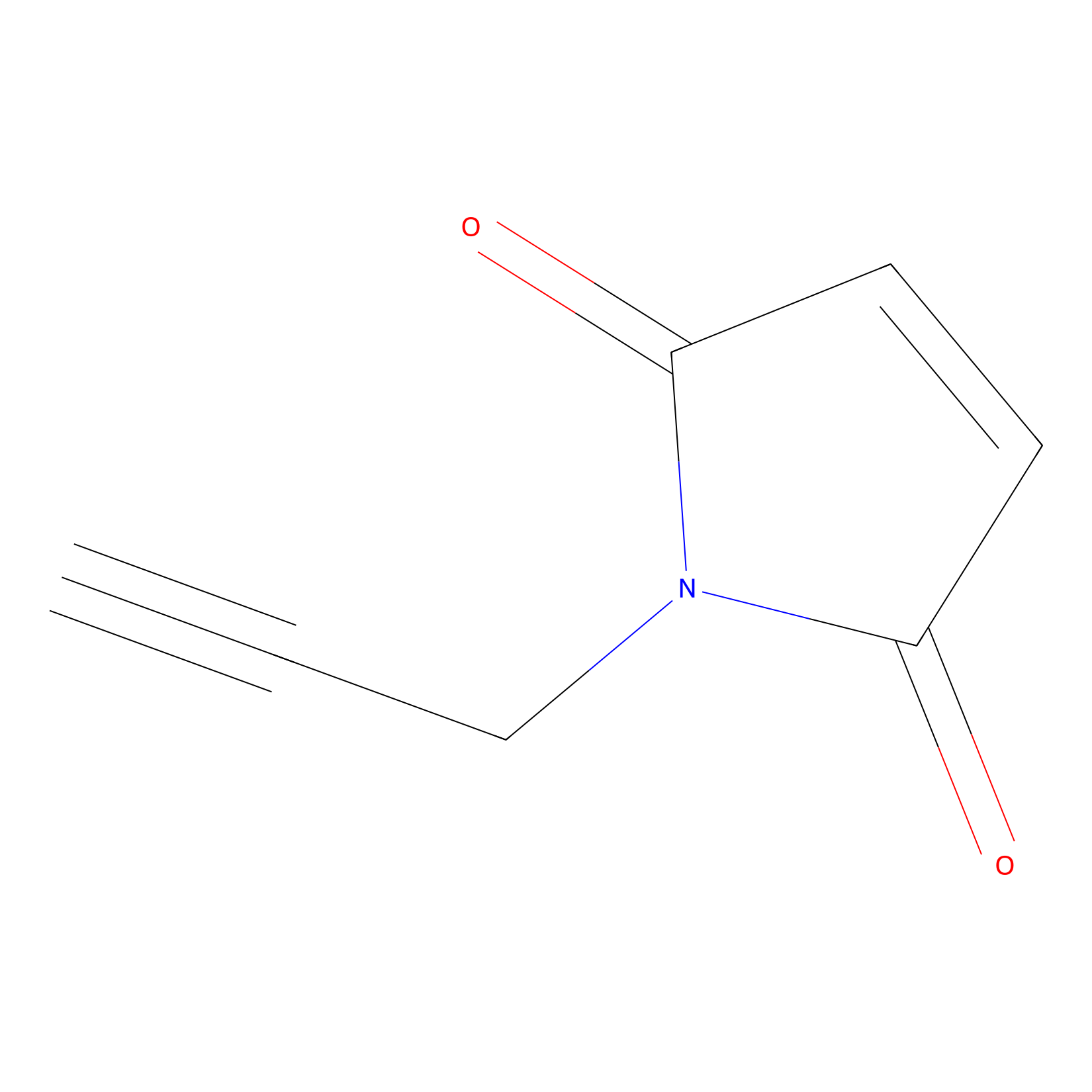 |
N.A. | LDD0016 | [10] | |
|
PPMS Probe Info |
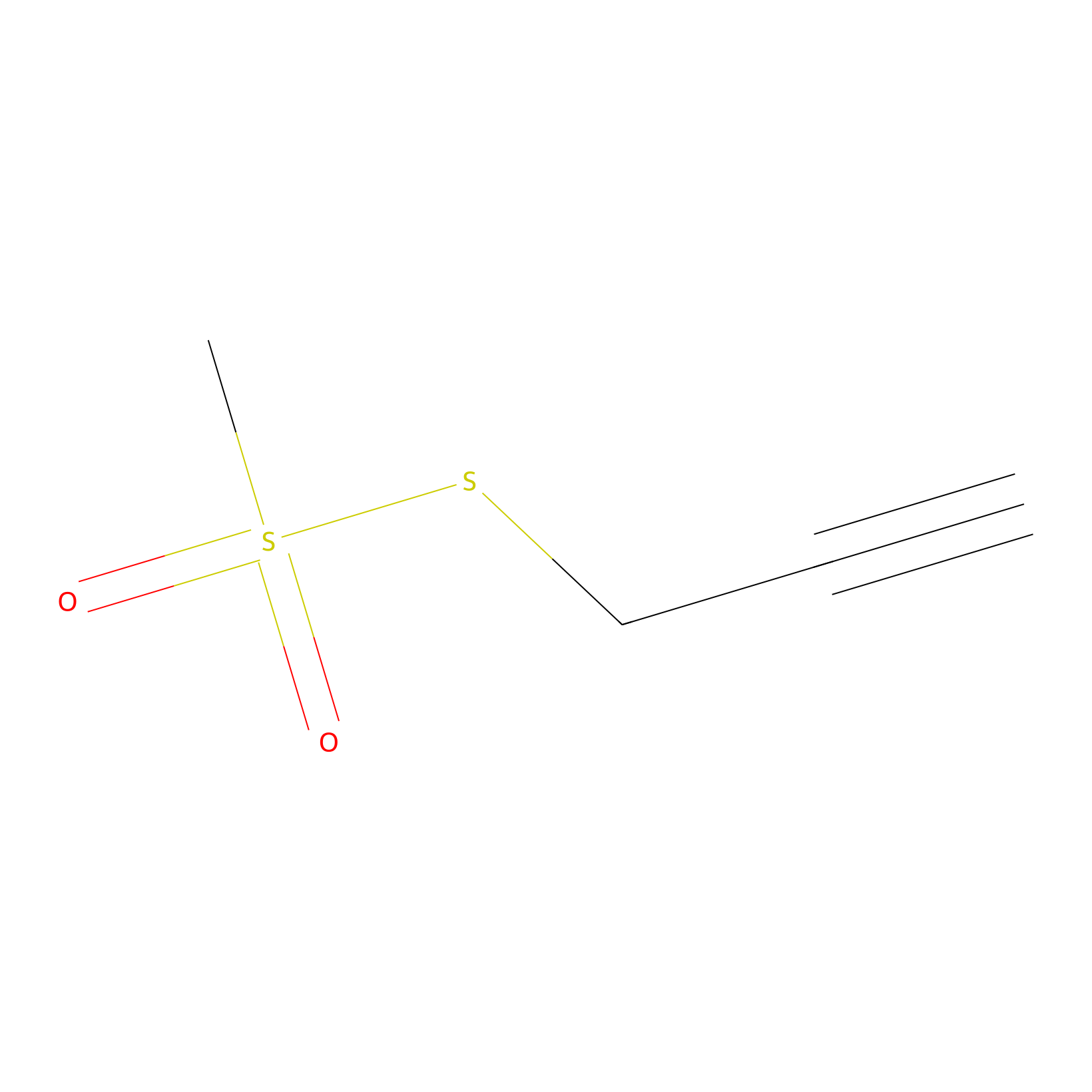 |
N.A. | LDD0008 | [10] | |
|
TFBX Probe Info |
 |
N.A. | LDD0148 | [9] | |
|
Phosphinate-6 Probe Info |
 |
N.A. | LDD0018 | [11] | |
|
Ox-W18 Probe Info |
 |
N.A. | LDD2175 | [12] | |
|
NAIA_5 Probe Info |
 |
C335(0.00); C133(0.00); C9(0.00) | LDD2223 | [13] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0116 | HHS-0101 | DM93 | Y316(0.74); Y317(0.81); Y24(0.86); Y235(1.01) | LDD0264 | [5] |
| LDCM0117 | HHS-0201 | DM93 | Y316(0.58); Y317(0.68); Y24(0.73); Y30(1.31) | LDD0265 | [5] |
| LDCM0118 | HHS-0301 | DM93 | Y317(0.70); Y316(0.70); Y235(0.76); Y24(0.80) | LDD0266 | [5] |
| LDCM0119 | HHS-0401 | DM93 | Y316(0.68); Y317(0.69); Y24(0.73); Y30(0.89) | LDD0267 | [5] |
| LDCM0120 | HHS-0701 | DM93 | Y316(0.45); Y317(0.60); Y24(0.64); Y30(0.73) | LDD0268 | [5] |
| LDCM0022 | KB02 | 42-MG-BA | C353(1.85); C151(2.74) | LDD2244 | [3] |
| LDCM0023 | KB03 | 42-MG-BA | C353(1.58); C151(2.63) | LDD2661 | [3] |
| LDCM0024 | KB05 | HMCB | C353(4.39); C151(6.91) | LDD3312 | [3] |
| LDCM0627 | NUDT7-COV-1 | HEK-293T | C9(0.59) | LDD2206 | [6] |
| LDCM0628 | OTUB2-COV-1 | HEK-293T | C9(0.71) | LDD2207 | [6] |
| LDCM0021 | THZ1 | HeLa S3 | C133(1.10) | LDD0460 | [4] |
References
