Details of the Target
General Information of Target
| Target ID | LDTP13380 | |||||
|---|---|---|---|---|---|---|
| Target Name | DNA helicase MCM8 (MCM8) | |||||
| Gene Name | MCM8 | |||||
| Gene ID | 84515 | |||||
| Synonyms |
C20orf154; DNA helicase MCM8; EC 3.6.4.12; Minichromosome maintenance 8 |
|||||
| 3D Structure | ||||||
| Sequence |
MLALRVARGSWGALRGAAWAPGTRPSKRRACWALLPPVPCCLGCLAERWRLRPAALGLRL
PGIGQRNHCSGAGKAAPRPAAGAGAAAEAPGGQWGPASTPSLYENPWTIPNMLSMTRIGL APVLGYLIIEEDFNIALGVFALAGLTDLLDGFIARNWANQRSALGSALDPLADKILISIL YVSLTYADLIPVPLTYMIISRDVMLIAAVFYVRYRTLPTPRTLAKYFNPCYATARLKPTF ISKVNTAVQLILVAASLAAPVFNYADSIYLQILWCFTAFTTAASAYSYYHYGRKTVQVIK D |
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
MCM family
|
|||||
| Subcellular location |
Nucleus
|
|||||
| Function |
Component of the MCM8-MCM9 complex, a complex involved in the repair of double-stranded DNA breaks (DBSs) and DNA interstrand cross-links (ICLs) by homologous recombination (HR). Required for DNA resection by the MRE11-RAD50-NBN/NBS1 (MRN) complex by recruiting the MRN complex to the repair site and by promoting the complex nuclease activity. Probably by regulating the localization of the MNR complex, indirectly regulates the recruitment of downstream effector RAD51 to DNA damage sites including DBSs and ICLs. The MCM8-MCM9 complex is dispensable for DNA replication and S phase progression. However, may play a non-essential for DNA replication: may be involved in the activation of the prereplicative complex (pre-RC) during G(1) phase by recruiting CDC6 to the origin recognition complex (ORC). Probably by regulating HR, plays a key role during gametogenesis. Stabilizes MCM9 protein.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
DBIA Probe Info |
 |
C588(5.49) | LDD3373 | [1] | |
|
IA-alkyne Probe Info |
 |
C155(0.00); C235(0.00) | LDD0165 | [2] | |
|
IPM Probe Info |
 |
C411(0.00); C235(0.00); C468(0.00) | LDD2156 | [3] | |
|
TFBX Probe Info |
 |
N.A. | LDD0148 | [4] | |
|
AOyne Probe Info |
 |
15.00 | LDD0443 | [5] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C153 Probe Info |
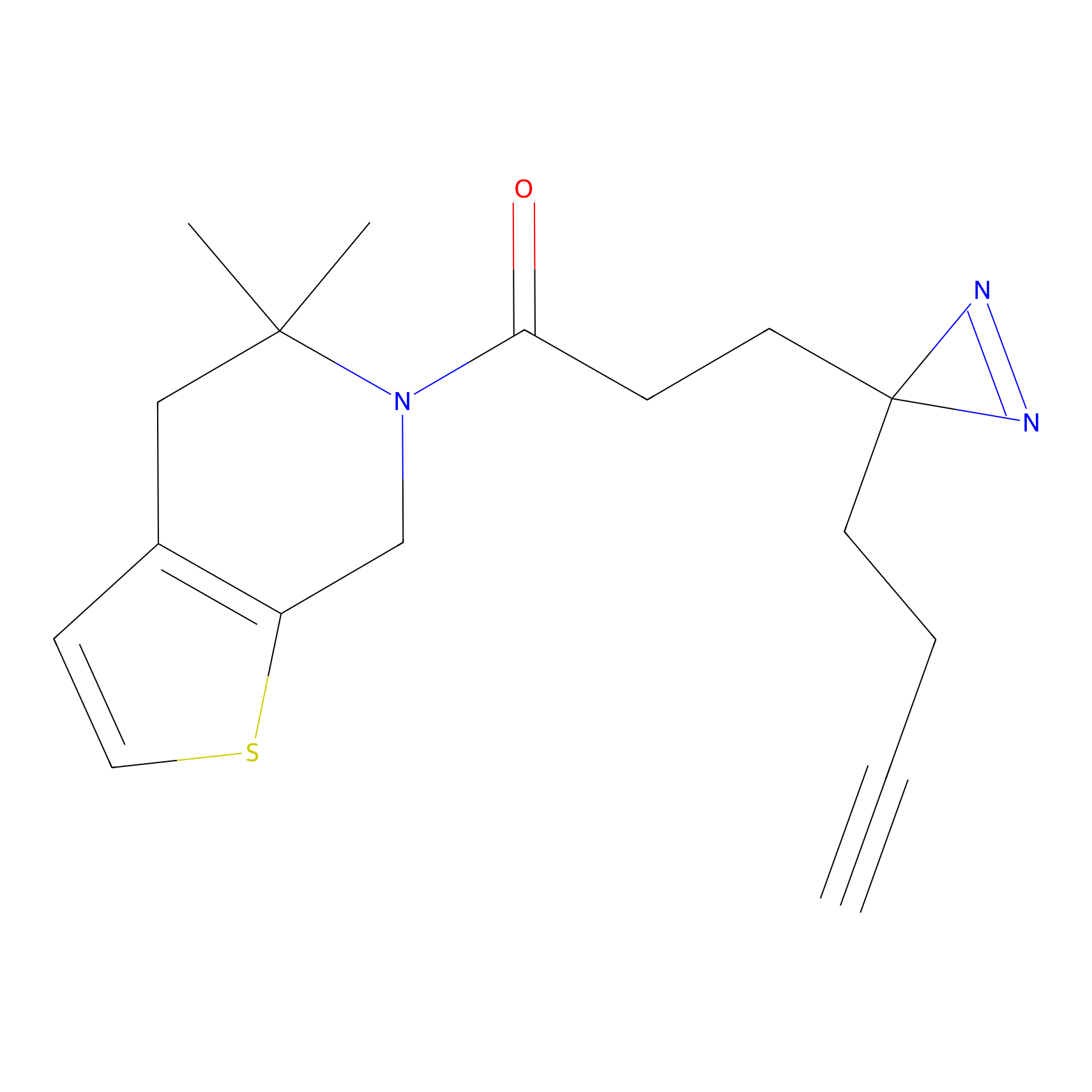 |
15.35 | LDD1834 | [6] | |
Competitor(s) Related to This Target
The Interaction Atlas With This Target
References
