Details of the Target
General Information of Target
Target Site Mutations in Different Cell Lines
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
TH211 Probe Info |
 |
Y58(15.82) | LDD0260 | [1] | |
|
AZ-9 Probe Info |
 |
E55(1.18) | LDD2208 | [2] | |
|
ONAyne Probe Info |
 |
K49(0.00); K71(0.00); K82(0.00) | LDD0273 | [3] | |
|
OPA-S-S-alkyne Probe Info |
 |
K49(3.39); K90(6.67) | LDD3494 | [4] | |
|
Probe 1 Probe Info |
 |
Y58(53.67) | LDD3495 | [5] | |
|
HHS-465 Probe Info |
 |
Y58(10.00) | LDD2237 | [6] | |
|
5E-2FA Probe Info |
 |
H80(0.00); H73(0.00) | LDD2235 | [7] | |
|
ATP probe Probe Info |
 |
K71(0.00); K72(0.00) | LDD0199 | [8] | |
|
m-APA Probe Info |
 |
H80(0.00); H74(0.00); H73(0.00) | LDD2233 | [7] | |
|
NHS Probe Info |
 |
N.A. | LDD0010 | [9] | |
|
SF Probe Info |
 |
K49(0.00); K82(0.00) | LDD0028 | [10] | |
|
STPyne Probe Info |
 |
K72(0.00); K82(0.00) | LDD0009 | [9] | |
|
Acrolein Probe Info |
 |
H74(0.00); H73(0.00) | LDD0217 | [11] | |
|
W1 Probe Info |
 |
N.A. | LDD0236 | [12] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C338 Probe Info |
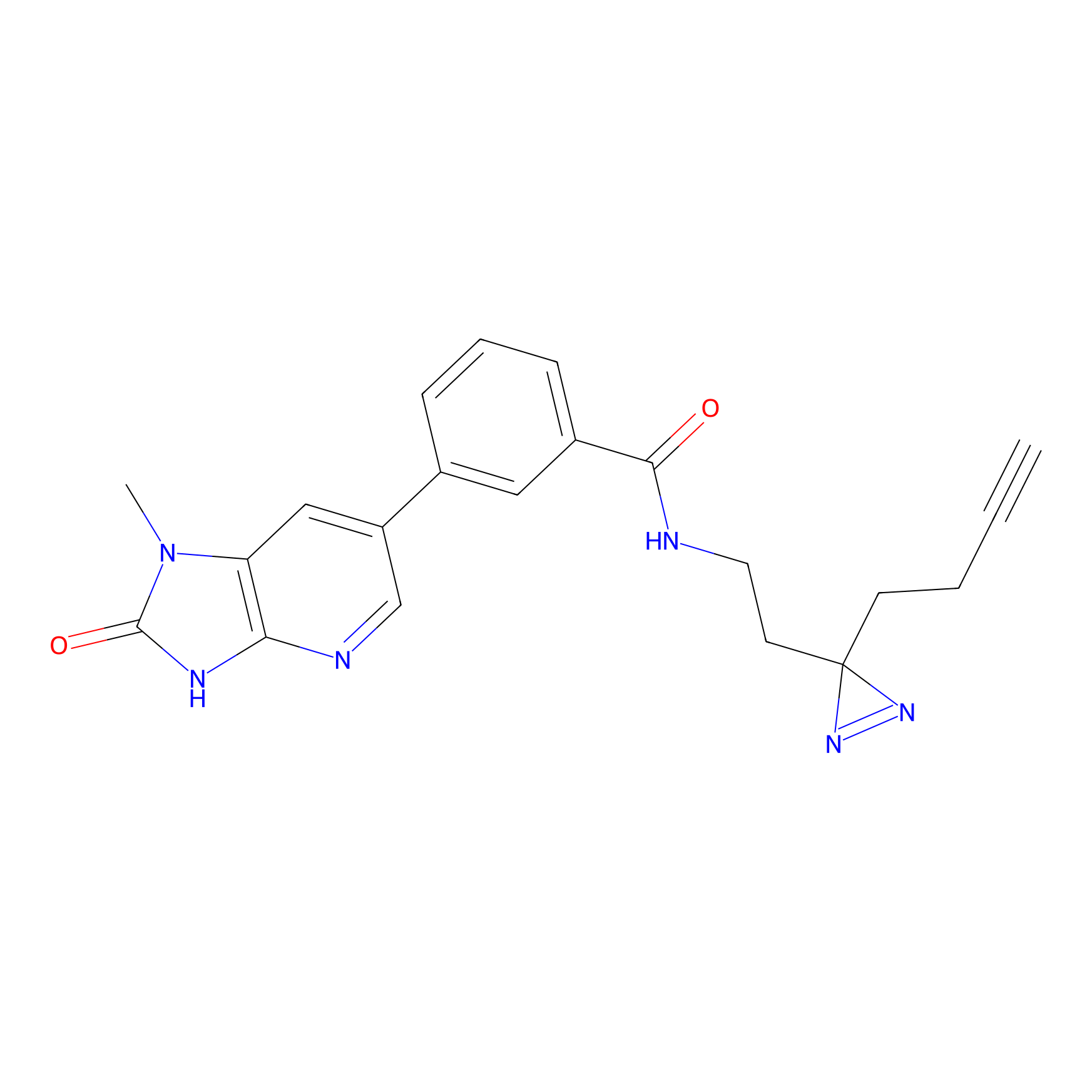 |
10.34 | LDD2001 | [13] | |
Competitor(s) Related to This Target
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| mRNA export factor GLE1 (GLE1) | GLE1 family | Q53GS7 | |||
| Nucleoporin p58/p45 (NUP58) | NUP58 family | Q9BVL2 | |||
Transcription factor
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Krueppel-like factor 11 (KLF11) | Sp1 C2H2-type zinc-finger protein family | O14901 | |||
GPCR
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Beta-2 adrenergic receptor (ADRB2) | G-protein coupled receptor 1 family | P07550 | |||
Cytokine and receptor
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Oncostatin-M-specific receptor subunit beta (OSMR) | Type I cytokine receptor family | Q99650 | |||
Other
References
