Details of the Target
General Information of Target
| Target ID | LDTP11705 | |||||
|---|---|---|---|---|---|---|
| Target Name | Bromodomain-containing protein 8 (BRD8) | |||||
| Gene Name | BRD8 | |||||
| Gene ID | 10902 | |||||
| Synonyms |
SMAP; SMAP2; Bromodomain-containing protein 8; Skeletal muscle abundant protein; Skeletal muscle abundant protein 2; Thyroid hormone receptor coactivating protein of 120 kDa; TrCP120; p120 |
|||||
| 3D Structure | ||||||
| Sequence |
MLAMDTCKHVGQLQLAQDHSSLNPQKWHCVDCNTTESIWACLSCSHVACGRYIEEHALKH
FQESSHPVALEVNEMYVFCYLCDDYVLNDNTTGDLKLLRRTLSAIKSQNYHCTTRSGRFL RSMGTGDDSYFLHDGAQSLLQSEDQLYTALWHRRRILMGKIFRTWFEQSPIGRKKQEEPF QEKIVVKREVKKRRQELEYQVKAELESMPPRKSLRLQGLAQSTIIEIVSVQVPAQTPASP AKDKVLSTSENEISQKVSDSSVKRRPIVTPGVTGLRNLGNTCYMNSVLQVLSHLLIFRQC FLKLDLNQWLAMTASEKTRSCKHPPVTDTVVYQMNECQEKDTGFVCSRQSSLSSGLSGGA SKGRKMELIQPKEPTSQYISLCHELHTLFQVMWSGKWALVSPFAMLHSVWRLIPAFRGYA QQDAQEFLCELLDKIQRELETTGTSLPALIPTSQRKLIKQVLNVVNNIFHGQLLSQVTCL ACDNKSNTIEPFWDLSLEFPERYQCSGKDIASQPCLVTEMLAKFTETEALEGKIYVCDQC NSKRRRFSSKPVVLTEAQKQLMICHLPQVLRLHLKRFRWSGRNNREKIGVHVGFEEILNM EPYCCRETLKSLRPECFIYDLSAVVMHHGKGFGSGHYTAYCYNSEGGFWVHCNDSKLSMC TMDEVCKAQAYILFYTQRVTENGHSKLLPPELLLGSQHPNEDADTSSNEILS |
|||||
| Target Bioclass |
Other
|
|||||
| Subcellular location |
Nucleus
|
|||||
| Function |
May act as a coactivator during transcriptional activation by hormone-activated nuclear receptors (NR). Isoform 2 stimulates transcriptional activation by AR/DHTR, ESR1/NR3A1, RXRA/NR2B1 and THRB/ERBA2. At least isoform 1 and isoform 2 are components of the NuA4 histone acetyltransferase (HAT) complex which is involved in transcriptional activation of select genes principally by acetylation of nucleosomal histones H4 and H2A. This modification may both alter nucleosome - DNA interactions and promote interaction of the modified histones with other proteins which positively regulate transcription. This complex may be required for the activation of transcriptional programs associated with oncogene and proto-oncogene mediated growth induction, tumor suppressor mediated growth arrest and replicative senescence, apoptosis, and DNA repair. NuA4 may also play a direct role in DNA repair when recruited to sites of DNA damage. Component of a SWR1-like complex that specifically mediates the removal of histone H2A.Z/H2AZ1 from the nucleosome.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
| ChEMBL ID | ||||||
Target Site Mutations in Different Cell Lines
| Cell line | Mutation details | Probe for labeling this protein in this cell | |||
|---|---|---|---|---|---|
| FTC133 | SNV: p.I495K | . | |||
| HCT116 | Deletion: p.R178GfsTer6 | . | |||
| HEC1 | SNV: p.A65T | DBIA Probe Info | |||
| HEC1B | SNV: p.A65T | . | |||
| HSC3 | Deletion: p.L1113QfsTer5 | . | |||
| MFE296 | Deletion: p.R178GfsTer6 | DBIA Probe Info | |||
| NCIH3122 | SNV: p.E657Ter | DBIA Probe Info | |||
| PEO1 | SNV: p.Y728H | . | |||
| YSCCC | SNV: p.S1158F | . | |||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
15.00 | LDD0402 | [1] | |
|
P8 Probe Info |
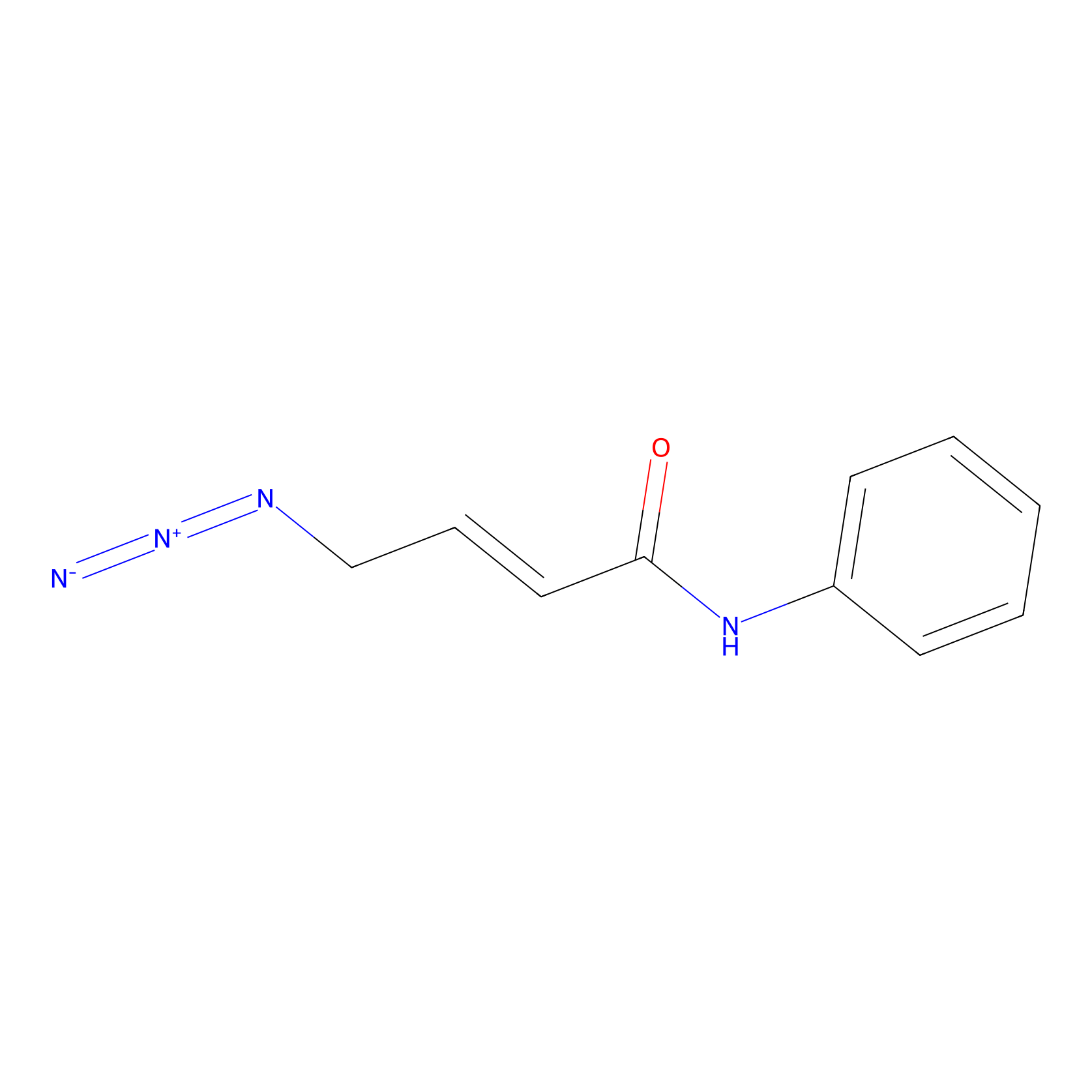 |
2.08 | LDD0455 | [2] | |
|
DBIA Probe Info |
 |
C26(2.75) | LDD3310 | [3] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0222 | [4] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0036 | [5] | |
|
Lodoacetamide azide Probe Info |
 |
N.A. | LDD0037 | [5] | |
|
IPM Probe Info |
 |
C142(0.00); C64(0.00) | LDD0147 | [6] | |
|
TFBX Probe Info |
 |
N.A. | LDD0148 | [6] | |
|
Phosphinate-6 Probe Info |
 |
N.A. | LDD0018 | [7] | |
|
Methacrolein Probe Info |
 |
N.A. | LDD0218 | [4] | |
|
W1 Probe Info |
 |
N.A. | LDD0236 | [8] | |
|
AOyne Probe Info |
 |
15.00 | LDD0443 | [9] | |
|
NAIA_5 Probe Info |
 |
C64(0.00); C26(0.00) | LDD2223 | [10] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0156 | Aniline | NCI-H1299 | 11.51 | LDD0403 | [1] |
| LDCM0108 | Chloroacetamide | HeLa | N.A. | LDD0222 | [4] |
| LDCM0632 | CL-Sc | Hep-G2 | C64(1.13); C64(0.95) | LDD2227 | [10] |
| LDCM0022 | KB02 | 22RV1 | C26(2.46) | LDD2243 | [3] |
| LDCM0023 | KB03 | 22RV1 | C26(3.74) | LDD2660 | [3] |
| LDCM0024 | KB05 | COLO792 | C26(2.75) | LDD3310 | [3] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Fibronectin type III and SPRY domain-containing protein 2 (FSD2) | . | A1L4K1 | |||
| Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 (PIN1) | . | Q13526 | |||
Other
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Enhancer of polycomb homolog 1 (EPC1) | Enhancer of polycomb family | Q9H2F5 | |||
| Protein Mis18-alpha (MIS18A) | Mis18 family | Q9NYP9 | |||
| Microspherule protein 1 (MCRS1) | . | Q96EZ8 | |||
References
