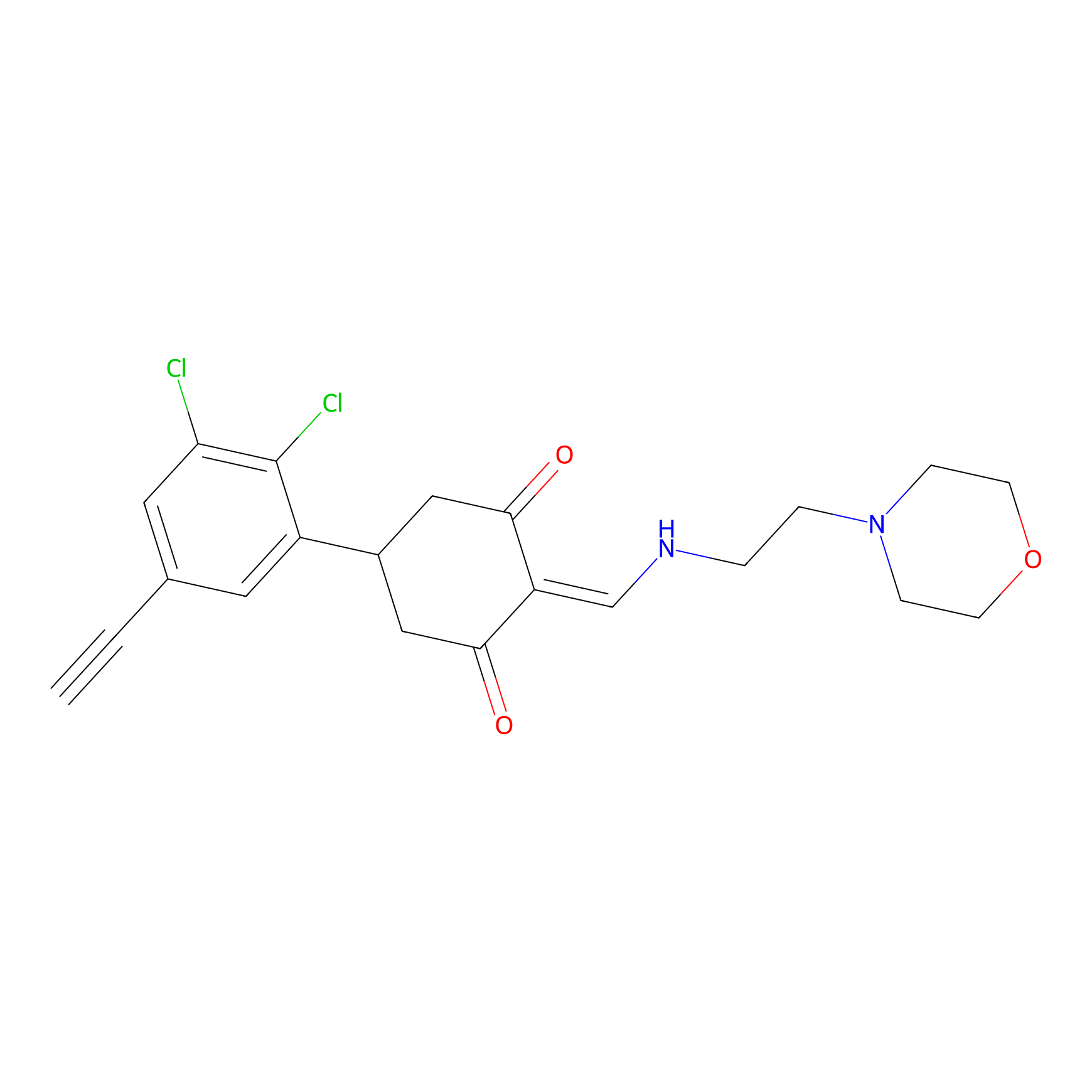
The Protein(s) Related To This Target
Enzyme
Click To Hide/Show 13 Protein(s) Interacting with
This Target
| Protein name |
Family |
Uniprot ID |
| Ubiquitin-like-conjugating enzyme ATG3 (ATG3) |
ATG3 family |
Q9NT62
|
| Trifunctional enzyme subunit alpha, mitochondrial (HADHA) |
Enoyl-CoA hydratase/isomerase family; 3-hydroxyacyl-CoA dehydrogenase family |
P40939
|
| Titin (TTN) |
CAMK Ser/Thr protein kinase family |
Q8WZ42
|
| Serine/threonine-protein kinase Nek9 (NEK9) |
NEK Ser/Thr protein kinase family |
Q8TD19
|
| Serine/threonine-protein kinase PINK1, mitochondrial (PINK1) |
Ser/Thr protein kinase family |
Q9BXM7
|
| Serine/threonine-protein kinase ULK1 (ULK1) |
Ser/Thr protein kinase family |
O75385
|
| Serine/threonine-protein kinase 3 (STK3) |
STE Ser/Thr protein kinase family |
Q13188
|
| Serine/threonine-protein kinase 4 (STK4) |
STE Ser/Thr protein kinase family |
Q13043
|
| Fibroblast growth factor receptor 3 (FGFR3) |
Tyr protein kinase family |
P22607
|
| NAD-dependent protein deacetylase sirtuin-1 (SIRT1) |
Sirtuin family |
Q96EB6
|
| Immunity-related GTPase family Q protein (IRGQ) |
IRG family |
Q8WZA9
|
| Ubiquitin-like modifier-activating enzyme 5 (UBA5) |
Ubiquitin-activating E1 family |
Q9GZZ9
|
| E3 ubiquitin-protein ligase NEDD4 (NEDD4) |
. |
P46934
|
Transporter and channel
Click To Hide/Show 11 Protein(s) Interacting with
This Target
| Protein name |
Family |
Uniprot ID |
| Amyloid-beta precursor protein (APP) |
APP family |
P05067
|
| Autophagy-related protein 13 (ATG13) |
ATG13 family |
O75143
|
| Autophagy-related protein 2 homolog A (ATG2A) |
ATG2 family |
Q2TAZ0
|
| Ubiquitin-like modifier-activating enzyme ATG7 (ATG7) |
ATG7 family |
O95352
|
| Caveolin-1 (CAV1) |
Caveolin family |
Q03135
|
| Clathrin interactor 1 (CLINT1) |
Epsin family |
Q14677
|
| Importin-5 (IPO5) |
Importin beta family |
O00410
|
| Cysteine protease ATG4B (ATG4B) |
Peptidase C54 family |
Q9Y4P1
|
| Autophagy-related protein 16-1 (ATG16L1) |
WD repeat ATG16 family |
Q676U5
|
| FYVE and coiled-coil domain-containing protein 1 (FYCO1) |
. |
Q9BQS8
|
| Starch-binding domain-containing protein 1 (STBD1) |
. |
O95210
|
Other
Click To Hide/Show 26 Protein(s) Interacting with
This Target
| Protein name |
Family |
Uniprot ID |
| Autophagy protein 5 (ATG5) |
ATG5 family |
Q9H1Y0
|
| Catenin beta-1 (CTNNB1) |
Beta-catenin family |
P35222
|
| FUN14 domain-containing protein 1 (FUNDC1) |
FUN14 family |
Q8IVP5
|
| Golgi reassembly-stacking protein 2 (GORASP2) |
GORASP family |
Q9H8Y8
|
| Progranulin (GRN) |
Granulin family |
P28799
|
| Heat shock 70 kDa protein 1A (HSPA1A) |
Heat shock protein 70 family |
P0DMV8
|
| Keratin, type II cytoskeletal 2 oral (KRT76) |
Intermediate filament family |
Q01546
|
| Lamin-B1 (LMNB1) |
Intermediate filament family |
P20700
|
| KxDL motif-containing protein 1 (KXD1) |
KXD1 family |
Q9BQD3
|
| Putative RNA-binding protein Luc7-like 2 (LUC7L2) |
Luc7 family |
Q9Y383
|
| LRP chaperone MESD (MESD) |
MESD family |
Q14696
|
| Protein NipSnap homolog 2 (NIPSNAP2) |
NipSnap family |
O75323
|
| Centriole and centriolar satellite protein OFD1 (OFD1) |
OFD1 family |
O75665
|
| Reticulophagy regulator 2 (RETREG2) |
RETREG family |
Q8NC44
|
| Kelch repeat and BTB domain-containing protein 6 (KBTBD6) |
. |
Q86V97
|
| Kelch repeat and BTB domain-containing protein 7 (KBTBD7) |
. |
Q8WVZ9
|
| Next to BRCA1 gene 1 protein (NBR1) |
. |
Q14596
|
| Ras association domain-containing protein 1 (RASSF1) |
. |
Q9NS23
|
| Ras association domain-containing protein 5 (RASSF5) |
. |
Q8WWW0
|
| Scaffold attachment factor B1 (SAFB) |
. |
Q15424
|
| Scaffold attachment factor B2 (SAFB2) |
. |
Q14151
|
| Sequestosome-1 (SQSTM1) |
. |
Q13501
|
| TBC1 domain family member 15 (TBC1D15) |
. |
Q8TC07
|
| TBC1 domain family member 2B (TBC1D2B) |
. |
Q9UPU7
|
| TNFAIP3-interacting protein 1 (TNIP1) |
. |
Q15025
|
| WD repeat and FYVE domain-containing protein 3 (WDFY3) |
. |
Q8IZQ1
|