Details of the Target
General Information of Target
| Target ID | LDTP10726 | |||||
|---|---|---|---|---|---|---|
| Target Name | Kinesin-like protein KIF20B (KIF20B) | |||||
| Gene Name | KIF20B | |||||
| Gene ID | 9585 | |||||
| Synonyms |
KRMP1; MPHOSPH1; Kinesin-like protein KIF20B; Cancer/testis antigen 90; CT90; Kinesin family member 20B; Kinesin-related motor interacting with PIN1; M-phase phosphoprotein 1; MPP1 |
|||||
| 3D Structure | ||||||
| Sequence |
MEEKRRRARVQGAWAAPVKSQAIAQPATTAKSHLHQKPGQTWKNKEHHLSDREFVFKEPQ
QVVRRAPEPRVIDREGVYEISLSPTGVSRVCLYPGFVDVKEADWILEQLCQDVPWKQRTG IREDITYQQPRLTAWYGELPYTYSRITMEPNPHWHPVLRTLKNRIEENTGHTFNSLLCNL YRNEKDSVDWHSDDEPSLGRCPIIASLSFGATRTFEMRKKPPPEENGDYTYVERVKIPLD HGTLLIMEGATQADWQHRVPKEYHSREPRVNLTFRTVYPDPRGAPW |
|||||
| Target Type |
Literature-reported
|
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
TRAFAC class myosin-kinesin ATPase superfamily, Kinesin family
|
|||||
| Subcellular location |
Nucleus
|
|||||
| Function |
Plus-end-directed motor enzyme that is required for completion of cytokinesis. Required for proper midbody organization and abscission in polarized cortical stem cells. Plays a role in the regulation of neuronal polarization by mediating the transport of specific cargos. Participates in the mobilization of SHTN1 and in the accumulation of PIP3 in the growth cone of primary hippocampal neurons in a tubulin and actin-dependent manner. In the developing telencephalon, cooperates with SHTN1 to promote both the transition from the multipolar to the bipolar stage and the radial migration of cortical neurons from the ventricular zone toward the superficial layer of the neocortex. Involved in cerebral cortex growth. Acts as an oncogene for promoting bladder cancer cells proliferation, apoptosis inhibition and carcinogenic progression.
|
|||||
| TTD ID | ||||||
| Uniprot ID | ||||||
| DrugMap ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
| ChEMBL ID | ||||||
Target Site Mutations in Different Cell Lines
| Cell line | Mutation details | Probe for labeling this protein in this cell | |||
|---|---|---|---|---|---|
| CAL72 | Deletion: p.N1328TfsTer7 | . | |||
| CALU6 | SNV: p.Q955K | DBIA Probe Info | |||
| DU145 | Deletion: p.V1353WfsTer15 | . | |||
| FUOV1 | Substitution: p.N756V | DBIA Probe Info | |||
| HCT15 | SNV: p.V1763A | . | |||
| HDQP1 | SNV: p.E1432Q | . | |||
| HEC1 | SNV: p.E277K | . | |||
| HEC1B | SNV: p.E277K | . | |||
| HSC3 | SNV: p.T1407I | . | |||
| HT115 | SNV: p.R574I | . | |||
| HUH1 | Substitution: p.N756V | . | |||
| ICC9 | Deletion: p.S930RfsTer10 | . | |||
| JVM3 | SNV: p.K1073R | . | |||
| KASUMI2 | Substitution: p.N756V | . | |||
| MCC13 | SNV: p.R1778P | . | |||
| MDAMB453 | SNV: p.D860H | . | |||
| MDST8 | SNV: p.S42F | DBIA Probe Info | |||
| MFE296 | Deletion: p.S1075VfsTer18 | . | |||
| MFE319 | Deletion: p.K592RfsTer4 | . | |||
| MOLT4 | SNV: p.L1225P | . | |||
| NOMO1 | SNV: p.T154S | . | |||
| REC1 | SNV: p.E590D | DBIA Probe Info | |||
| REH | SNV: p.R328K | . | |||
| RT112 | SNV: p.E225Q | . | |||
| SHP77 | SNV: p.M1467I | . | |||
| SKHEP1 | SNV: p.K296E | . | |||
| SNU1 | Insertion: p.S1075KfsTer5 | DBIA Probe Info | |||
| SW1271 | SNV: p.R1677I | . | |||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
15.00 | LDD0402 | [1] | |
|
Jackson_1 Probe Info |
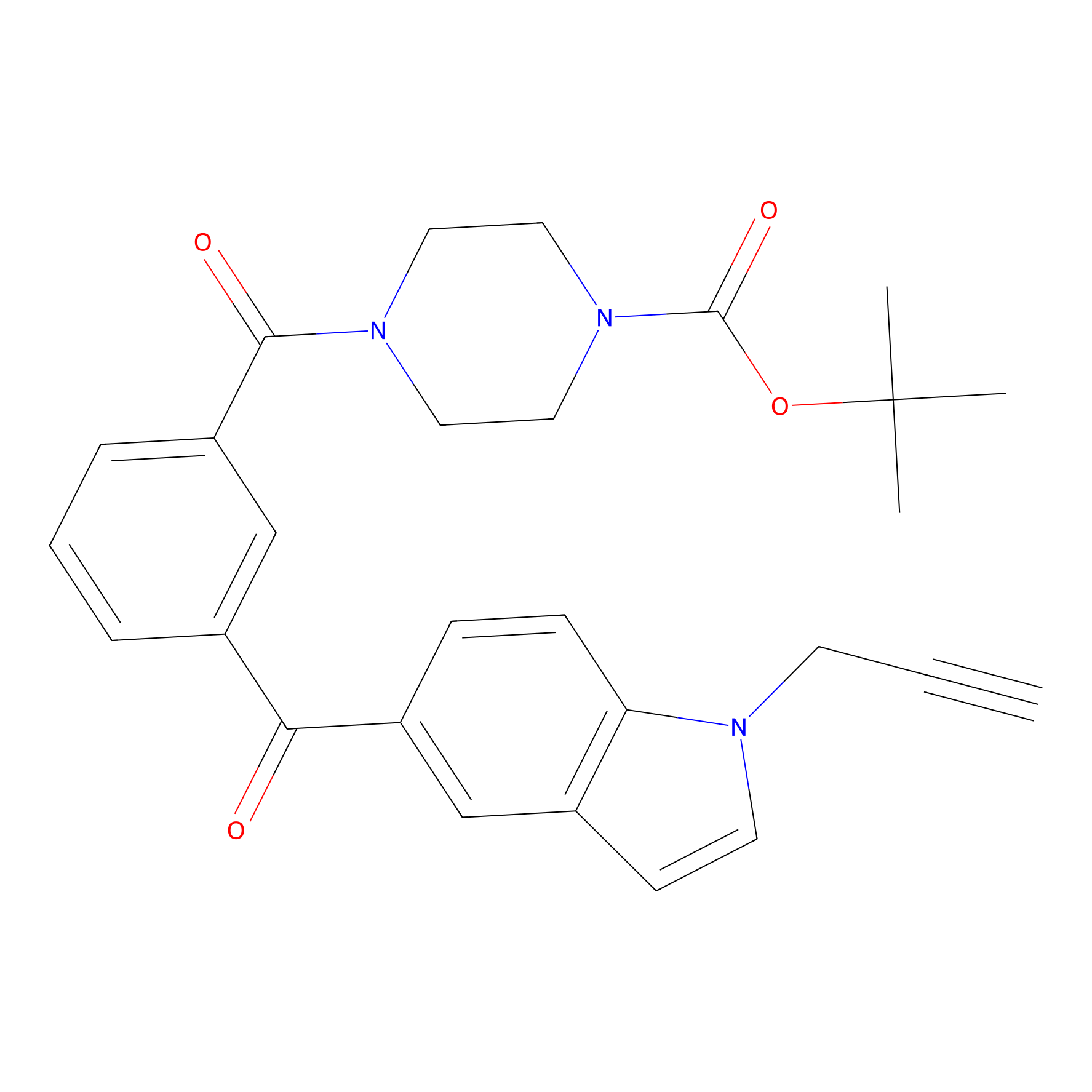 |
20.00 | LDD0122 | [2] | |
|
DBIA Probe Info |
 |
C480(3.87); C1210(1.24) | LDD3342 | [3] | |
|
NAIA_5 Probe Info |
 |
C1158(1.06) | LDD2227 | [4] | |
|
CY-1 Probe Info |
 |
N.A. | LDD0246 | [5] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0163 | [6] | |
|
Lodoacetamide azide Probe Info |
 |
N.A. | LDD0037 | [7] | |
|
ENE Probe Info |
 |
N.A. | LDD0006 | [8] | |
|
IPM Probe Info |
 |
N.A. | LDD0005 | [8] | |
|
NPM Probe Info |
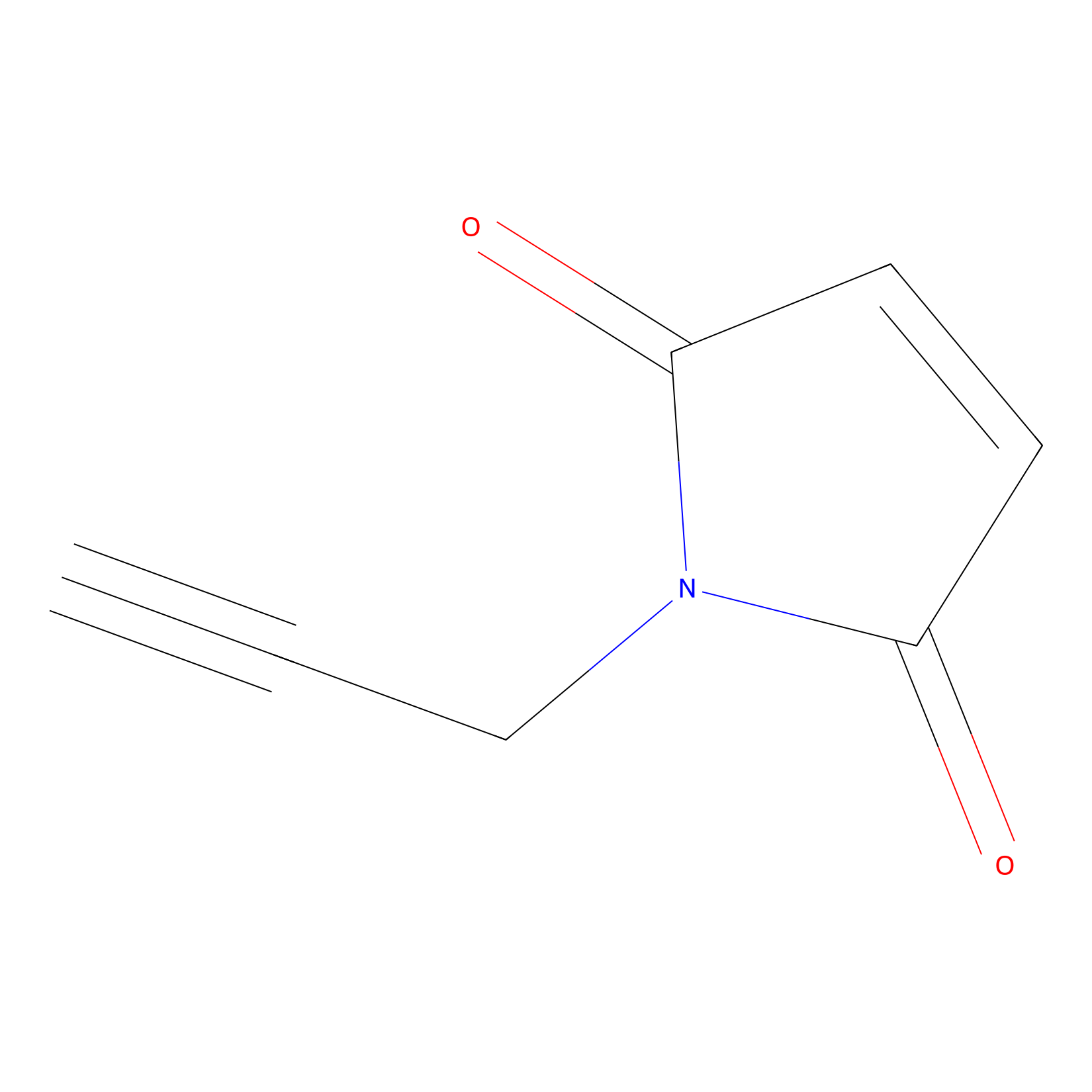 |
N.A. | LDD0016 | [8] | |
|
VSF Probe Info |
 |
N.A. | LDD0007 | [8] | |
|
AOyne Probe Info |
 |
14.20 | LDD0443 | [9] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0632 | CL-Sc | Hep-G2 | C1158(1.06) | LDD2227 | [4] |
| LDCM0634 | CY-0357 | Hep-G2 | C1642(50.00) | LDD2228 | [4] |
| LDCM0022 | KB02 | HEK-293T | C1642(0.98); C96(0.87); C480(0.89) | LDD1492 | [10] |
| LDCM0023 | KB03 | HEK-293T | C1642(0.97); C96(1.01); C480(1.03) | LDD1497 | [10] |
| LDCM0024 | KB05 | NCI-H1155 | C480(3.87); C1210(1.24) | LDD3342 | [3] |
References
