Details of the Target
General Information of Target
| Target ID | LDTP08239 | |||||
|---|---|---|---|---|---|---|
| Target Name | Zinc finger homeobox protein 4 (ZFHX4) | |||||
| Gene Name | ZFHX4 | |||||
| Gene ID | 79776 | |||||
| Synonyms |
Zinc finger homeobox protein 4; Zinc finger homeodomain protein 4; ZFH-4 |
|||||
| 3D Structure | ||||||
| Sequence |
METCDSPPISRQENGQSTSKLCGTTQLDNEVPEKVAGMEPDRENSSTDDNLKTDERKSEA
LLGFSVENAAATQVTSAKEIPCNECATSFPSLQKYMEHHCPNARLPVLKDDNESEISELE DSDVENLTGEIVYQPDGSAYIIEDSKESGQNAQTGANSKLFSTAMFLDSLASAGEKSDQS ASAPMSFYPQIINTFHIASSLGKPFTADQAFPNTSALAGVGPVLHSFRVYDLRHKREKDY LTSDGSAKNSCVSKDVPNNVDLSKFDGCVSDGKRKPVLMCFLCKLSFGYIRSFVTHAVHD HRMTLNDEEQKLLSNKCVSAIIQGIGKDKEPLISFLEPKKSTSVYPHFSTTNLIGPDPTF RGLWSAFHVENGDSLPAGFAFLKGSASTSSSAEQPLGITQMPKAEVNLGGLSSLVVNTPI TSVSLSHSSSESSKMSESKDQENNCERPKESNVLHPNGECPVKSEPTEPGDEDEEDAYSN ELDDEEVLGELTDSIGNKDFPLLNQSISPLSSSVLKFIEKGTSSSSATVSDDTEKKKQTA AVRASGSVASNYGISGKDFADASASKDSATAAHPSEIARGDEDSSATPHQHGFTPSTPGT PGPGGDGSPGSGIECPKCDTVLGSSRSLGGHMTMMHSRNSCKTLKCPKCNWHYKYQQTLE AHMKEKHPEPGGSCVYCKTGQPHPRLARGESYTCGYKPFRCEVCNYSTTTKGNLSIHMQS DKHLNNVQNLQNGNGEQVFGHSAPAPNTSLSGCGTPSPSKPKQKPTWRCEVCDYETNVAR NLRIHMTSEKHMHNMMLLQQNMKQIQHNLHLGLAPAEAELYQYYLAQNIGLTGMKLENPA DPQLMINPFQLDPATAAALAPGLGELSPYISDPALKLFQCAVCNKFTSDSLEALSVHVSS ERSLPEEEWRAVIGDIYQCKLCNYNTQLKANFQLHCKTDKHMQKYQLVAHIKEGGKSNEW RLKCIAIGNPVHLKCNACDYYTNSVDKLRLHTTNHRHEAALKLYKHLQKQEGAVNPESCY YYCAVCDYTTKVKLNLVQHVRSVKHQQTEGLRKLQLHQQGLAPEEDNLSEIFFVKDCPPN ELETASLGARTCDDDLTEQHEEAEGAIKPTAVAEDDEKDTSERDNSEGKNSNKDSVSVAG GTQPLLLAKEEDVATKRSKPTEDNKFCHEQFYQCPYCNYNSRDQSRIQMHVLSQHSVQPV ICCPLCQDVLSNKMHLQLHLTHLHSVSPDCVEKLLMTVPVPDVMMPNSMLLPAAASEKSE RDTPAAVTAEGSGKYSGESPMDDKSMAGLEDSKANVEVKNEEQKPTKEPLEVSEWNKNSS KDVKIPDTLQDQLNEQQKRQPLSVSDRHVYKYRCNHCSLAFKTMQKLQIHSQYHAIRAAT MCNLCQRSFRTFQALKKHLEAGHPELSEAELQQLYASLPVNGELWAESETMSQDDHGLEQ EMEREYEVDHEGKASPVGSDSSSIPDDMGSEPKRTLPFRKGPNFTMEKFLDPSRPYKCTV CKESFTQKNILLVHYNSVSHLHKLKKVLQEASSPVPQETNSNTDNKPYKCSICNVAYSQS STLEIHMRSVLHQTKARAAKLEPSGHVAGGHSIAANVNSPGQGMLDSMSLAAVNSKDTHL DAKELNKKQTPDLISAQPAHHPPQSPAQIQMQLQHELQQQAAFFQPQFLNPAFLPHFPMT PEALLQFQQPQFLFPFYIPGTEFSLGPDLGLPGSATFGMPGMTGMAGSLLEDLKQQIQTQ HHVGQTQLQILQQQAQQYQATQPQLQPQKQQQQPPPPQQQQQQQASKLLKQEQSNIVSAD CQIMKDVPSYKEAEDISEKPEKPKQEFISEGEGLKEGKDTKKQKSLEPSIPPPRIASGAR GNAAKALLENFGFELVIQYNENRQKVQKKGKSGEGENTDKLECGTCGKLFSNVLILKSHQ EHVHGQFFPYAALEKFARQYREAYDKLYPISPSSPETPPPPPPPPPLPPAPPQPSSMGPV KIPNTVSTPLQAPPPTPPPPPPPPPPPPPPPPPPPPSAPPQVQLPVSLDLPLFPSIMMQP VQHPALPPQLALQLPQMDALSADLTQLCQQQLGLDPNFLRHSQFKRPRTRITDDQLKILR AYFDINNSPSEEQIQEMAEKSGLSQKVIKHWFRNTLFKERQRNKDSPYNFSNPPITVLED IRIDPQPTSLEHYKSDASFSKRSSRTRFTDYQLRVLQDFFDTNAYPKDDEIEQLSTVLNL PTRVIVVWFQNARQKARKSYENQAETKDNEKRELTNERYIRTSNMQYQCKKCNVVFPRIF DLITHQKKQCYKDEDDDAQDESQTEDSMDATDQVVYKHCTVSGQTDAAKNAAAPAASSGS GTSTPLIPSPKPEPEKTSPKPEYPAEKPKQSDPSPPSQGTKPALPLASTSSDPPQASTAQ PQPQPQPPKQPQLIGRPPSASQTPVPSSPLQISMTSLQNSLPPQLLQYQCDQCTVAFPTL ELWQEHQHMHFLAAQNQFLHSPFLERPMDMPYMIFDPNNPLMTGQLLGSSLTQMPPQASS SHTTAPTTVAASLKRKLDDKEDNNCSEKEGGNSGEDQHRDKRLRTTITPEQLEILYEKYL LDSNPTRKMLDHIAREVGLKKRVVQVWFQNTRARERKGQFRAVGPAQSHKRCPFCRALFK AKSALESHIRSRHWNEGKQAGYSLPPSPLISTEDGGESPQKYIYFDYPSLPLTKIDLSSE NELASTVSTPVSKTAELSPKNLLSPSSFKAECSEDVENLNAPPAEAGYDQNKTDFDETSS INTAISDATTGDEGNTEMESTTGSSGDVKPALSPKEPKTLDTLPKPATTPTTEVCDDKFL FSLTSPSIHFNDKDGDHDQSFYITDDPDDNADRSETSSIADPSSPNPFGSSNPFKSKSND RPGHKRFRTQMSNLQLKVLKACFSDYRTPTMQECEMLGNEIGLPKRVVQVWFQNARAKEK KFKINIGKPFMINQGGTEGTKPECTLCGVKYSARLSIRDHIFSKQHISKVRETVGSQLDR EKDYLAPTTVRQLMAQQELDRIKKASDVLGLTVQQPGMMDSSSLHGISLPTAYPGLPGLP PVLLPGMNGPSSLPGFPQNSNISAGMLGFPTSATSSPALSLSSAPTKPLLQTPPPPPPPP PPPPSSSLSGQQTEQQNKESEKKQTKPNKVKKIKEEELEATKPEKHPKKEEKISSALSVL GKVVGETHVDPIQLQALQNAIAGDPASFIGGQFLPYFIPGFASYFTPQLPGTVQGGYFPP VCGMESLFPYGPTMPQTLAGLSPGALLQQYQQYQQNLQESLQKQQKQQQEQQQKPVQAKT SKVESDQPQNSNDASETKEDKSTATESTKEEPQLESKSADFSDTYVVPFVKYEFICRKCQ MMFTDEDAAVNHQKSFCYFGQPLIDPQETVLRVPVSKYQCLACDVAISGNEALSQHLQSS LHKEKTIKQAMRNAKEHVRLLPHSVCSPNPNTTSTSQSAASSNNTYPHLSCFSMKSWPNI LFQASARRAASPPSSPPSLSLPSTVTSSLCSTSGVQTSLPTESCSDESDSELSQKLEDLD NSLEVKAKPASGLDGNFNSIRMDMFSV |
|||||
| Target Bioclass |
Transcription factor
|
|||||
| Family |
Krueppel C2H2-type zinc-finger protein family
|
|||||
| Subcellular location |
Nucleus
|
|||||
| Function | May play a role in neural and muscle differentiation. May be involved in transcriptional regulation. | |||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
14.07 | LDD0402 | [2] | |
|
DBIA Probe Info |
 |
C445(1.04) | LDD3310 | [3] | |
|
AHL-Pu-1 Probe Info |
 |
C460(9.84) | LDD0168 | [4] | |
|
IA-alkyne Probe Info |
 |
C2319(0.00); C2902(0.00); C445(0.00) | LDD0165 | [5] | |
|
IPM Probe Info |
 |
N.A. | LDD0005 | [6] | |
|
NPM Probe Info |
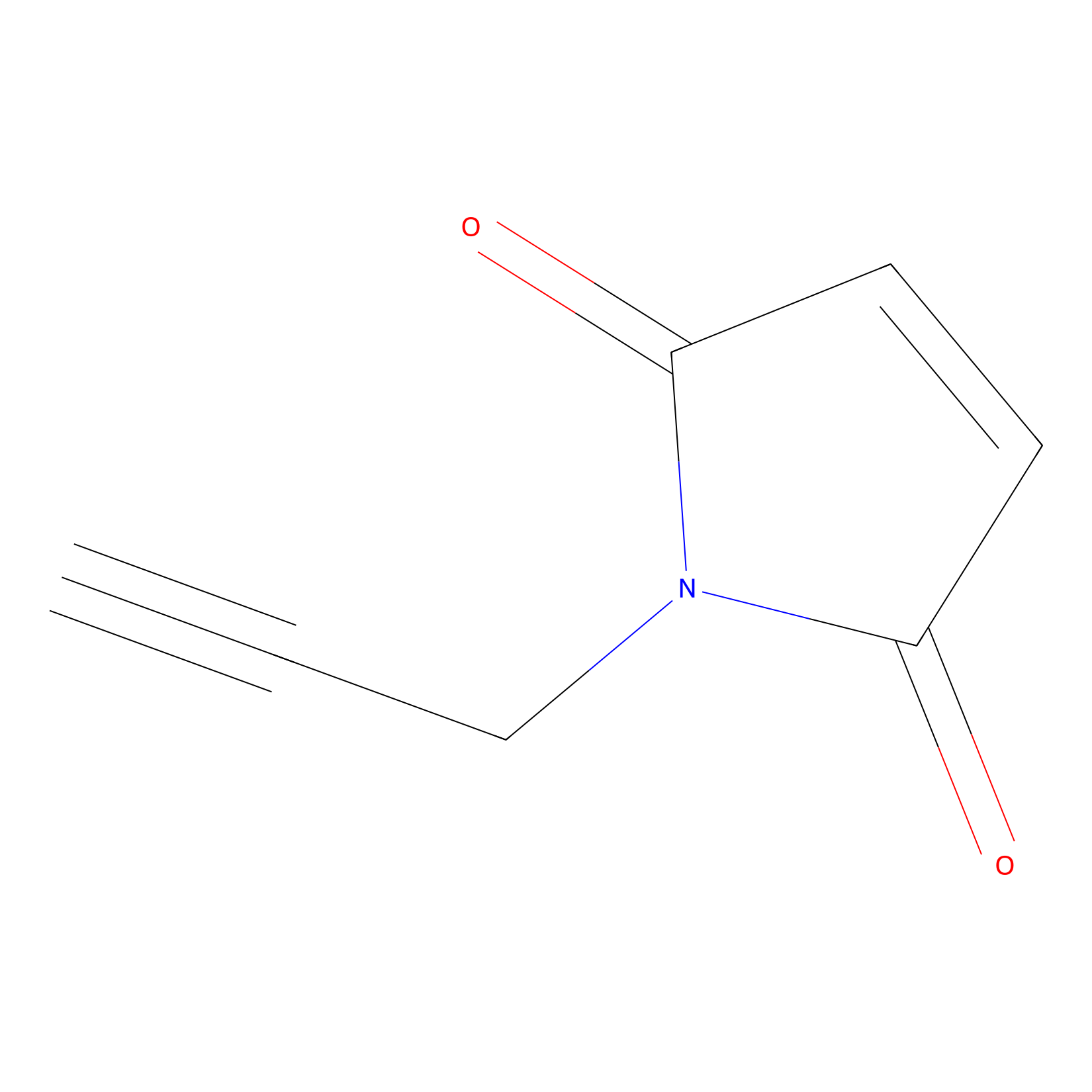 |
N.A. | LDD0016 | [6] | |
|
VSF Probe Info |
 |
N.A. | LDD0007 | [6] | |
|
Phosphinate-6 Probe Info |
 |
C100(0.00); C964(0.00) | LDD0018 | [7] | |
|
AOyne Probe Info |
 |
11.60 | LDD0443 | [8] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0025 | 4SU-RNA | HEK-293T | C460(9.84) | LDD0168 | [4] |
| LDCM0026 | 4SU-RNA+native RNA | HEK-293T | C460(6.49) | LDD0169 | [4] |
| LDCM0156 | Aniline | NCI-H1299 | 12.22 | LDD0403 | [2] |
| LDCM0022 | KB02 | 786-O | C645(1.16); C100(1.31) | LDD2247 | [3] |
| LDCM0023 | KB03 | 786-O | C645(1.31); C100(1.38) | LDD2664 | [3] |
| LDCM0024 | KB05 | COLO792 | C445(1.04) | LDD3310 | [3] |
References
