Details of the Target
General Information of Target
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
TG42 Probe Info |
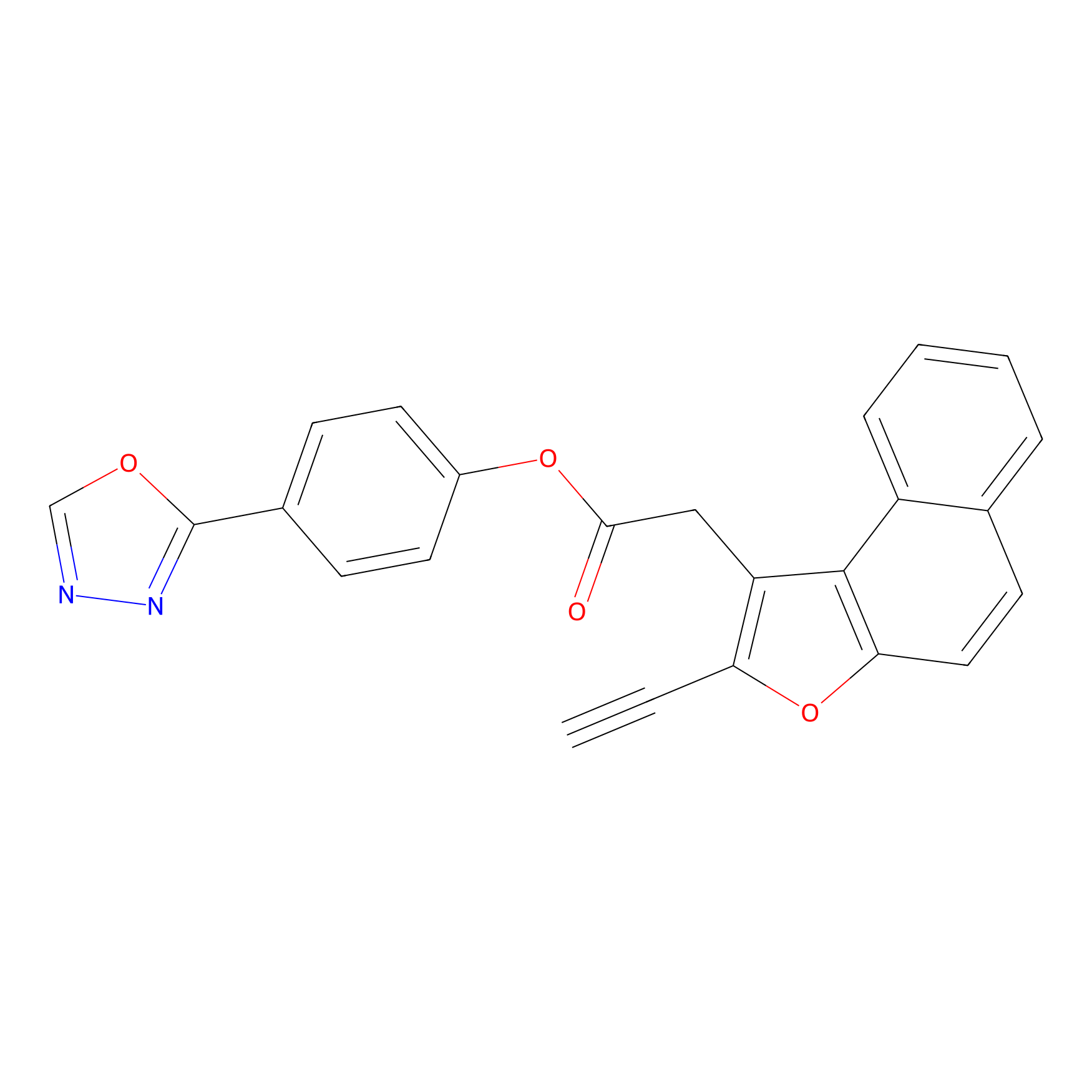 |
26.72 | LDD0326 | [1] | |
|
IPM Probe Info |
 |
N.A. | LDD0241 | [2] | |
|
OPA-S-S-alkyne Probe Info |
 |
K482(1.94); K531(2.47) | LDD3494 | [3] | |
|
Probe 1 Probe Info |
 |
Y301(12.53); Y342(16.45) | LDD3495 | [4] | |
|
DBIA Probe Info |
 |
C522(3.57); C337(1.85); C172(2.38) | LDD3312 | [5] | |
|
5E-2FA Probe Info |
 |
N.A. | LDD2235 | [6] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0163 | [7] | |
|
NAIA_5 Probe Info |
 |
C522(0.00); C473(0.00); C337(0.00) | LDD2223 | [8] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0020 | ARS-1620 | HCC44 | C628(1.24) | LDD2171 | [9] |
| LDCM0022 | KB02 | 22RV1 | C172(1.47) | LDD2243 | [5] |
| LDCM0023 | KB03 | 22RV1 | C172(1.65) | LDD2660 | [5] |
| LDCM0024 | KB05 | HMCB | C522(3.57); C337(1.85); C172(2.38) | LDD3312 | [5] |
| LDCM0021 | THZ1 | HCT 116 | C628(1.24) | LDD2173 | [9] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| E3 ubiquitin-protein ligase TRIM7 (TRIM7) | TRIM/RBCC family | Q9C029 | |||
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| MyoD family inhibitor (MDFI) | MDFI family | Q99750 | |||
| Kelch-like protein 2 (KLHL2) | . | O95198 | |||
Other
References
