Details of the Target
General Information of Target
| Target ID | LDTP03895 | |||||
|---|---|---|---|---|---|---|
| Target Name | All-trans-retinol dehydrogenase [NAD(+)] ADH7 (ADH7) | |||||
| Gene Name | ADH7 | |||||
| Gene ID | 131 | |||||
| Synonyms |
All-trans-retinol dehydrogenase [NAD(+)] ADH7; EC 1.1.1.105; Alcohol dehydrogenase class 4 mu/sigma chain; EC 1.1.1.1; Alcohol dehydrogenase class IV mu/sigma chain; Gastric alcohol dehydrogenase; Omega-hydroxydecanoate dehydrogenase ADH7; EC 1.1.1.66; Retinol dehydrogenase
|
|||||
| 3D Structure | ||||||
| Sequence |
MFAEIQIQDKDRMGTAGKVIKCKAAVLWEQKQPFSIEEIEVAPPKTKEVRIKILATGICR
TDDHVIKGTMVSKFPVIVGHEATGIVESIGEGVTTVKPGDKVIPLFLPQCRECNACRNPD GNLCIRSDITGRGVLADGTTRFTCKGKPVHHFMNTSTFTEYTVVDESSVAKIDDAAPPEK VCLIGCGFSTGYGAAVKTGKVKPGSTCVVFGLGGVGLSVIMGCKSAGASRIIGIDLNKDK FEKAMAVGATECISPKDSTKPISEVLSEMTGNNVGYTFEVIGHLETMIDALASCHMNYGT SVVVGVPPSAKMLTYDPMLLFTGRTWKGCVFGGLKSRDDVPKLVTEFLAKKFDLDQLITH VLPFKKISEGFELLNSGQSIRTVLTF |
|||||
| Target Type |
Literature-reported
|
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
Zinc-containing alcohol dehydrogenase family, Class-IV subfamily
|
|||||
| Subcellular location |
Cytoplasm
|
|||||
| Function |
Catalyzes the NAD-dependent oxidation of all-trans-retinol, alcohol, and omega-hydroxy fatty acids and their derivatives. Oxidizes preferentially all trans-retinol, all-trans-4-hydroxyretinol, 9-cis-retinol, 2-hexenol, and long chain omega-hydroxy fatty acids such as juniperic acid. In vitro can also catalyzes the NADH-dependent reduction of all-trans-retinal and aldehydes and their derivatives. Reduces preferentially all trans-retinal, all-trans-4-oxoretinal and hexanal. Catalyzes in the oxidative direction with higher efficiency. Therefore may participate in retinoid metabolism, fatty acid omega-oxidation, and elimination of cytotoxic aldehydes produced by lipid peroxidation.
|
|||||
| TTD ID | ||||||
| Uniprot ID | ||||||
| DrugMap ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
| ChEMBL ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
DBIA Probe Info |
 |
C118(1.55) | LDD3352 | [1] | |
|
P26 Probe Info |
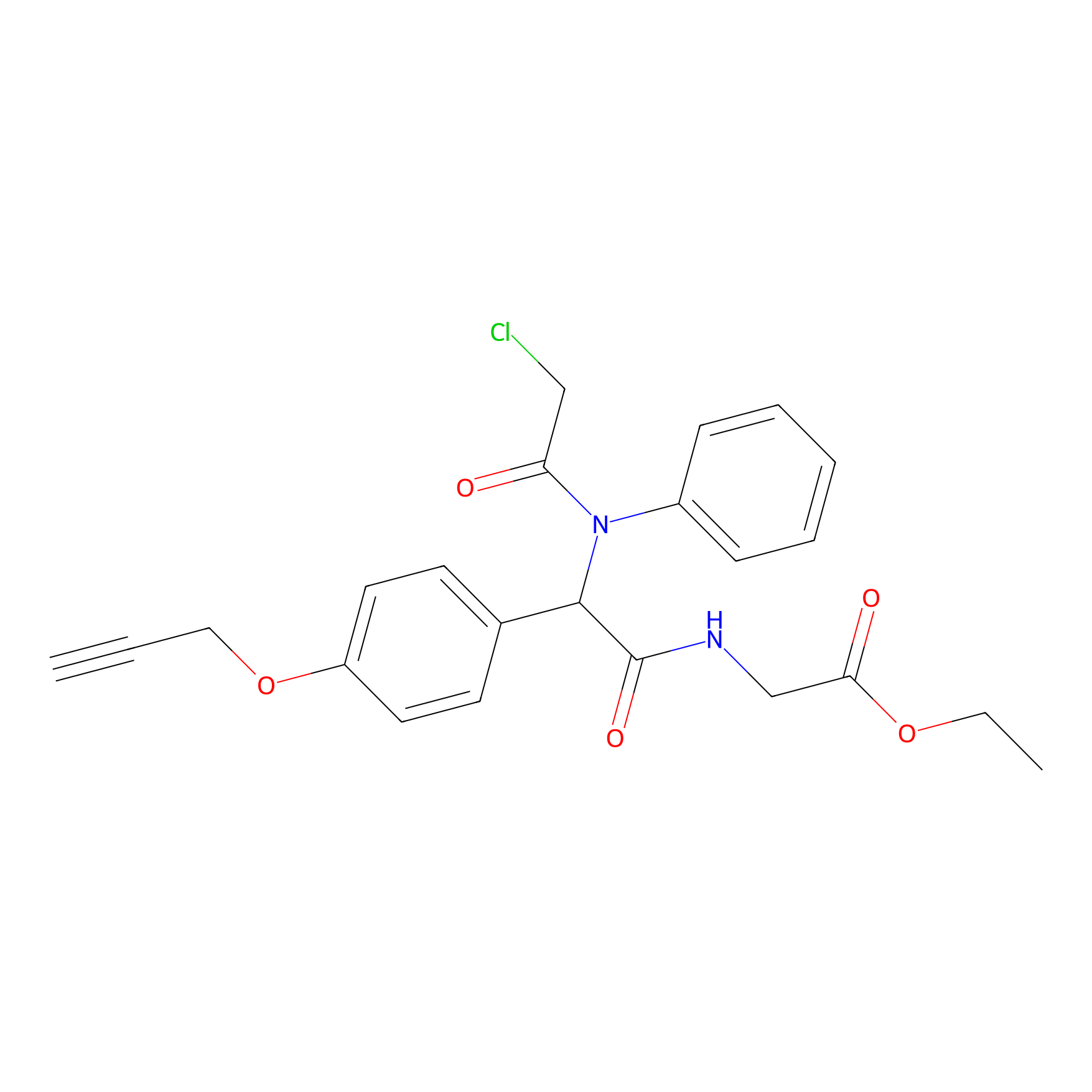 |
2.73 | LDD0409 | [2] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0222 | [3] | |
|
TFBX Probe Info |
 |
N.A. | LDD0148 | [4] | |
Competitor(s) Related to This Target
The Interaction Atlas With This Target
References
