Details of the Target
General Information of Target
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
TH211 Probe Info |
 |
Y61(7.95); Y54(7.41) | LDD0257 | [1] | |
|
STPyne Probe Info |
 |
K116(10.00); K126(7.59) | LDD0277 | [2] | |
|
ONAyne Probe Info |
 |
N.A. | LDD0273 | [2] | |
|
OPA-S-S-alkyne Probe Info |
 |
K16(1.12); K14(1.12) | LDD3494 | [3] | |
|
5E-2FA Probe Info |
 |
N.A. | LDD2235 | [4] | |
|
1c-yne Probe Info |
 |
N.A. | LDD0228 | [5] | |
|
HHS-465 Probe Info |
 |
N.A. | LDD2240 | [6] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
FFF probe3 Probe Info |
 |
5.54 | LDD0464 | [7] | |
|
STS-1 Probe Info |
 |
N.A. | LDD0136 | [8] | |
|
Photocelecoxib Probe Info |
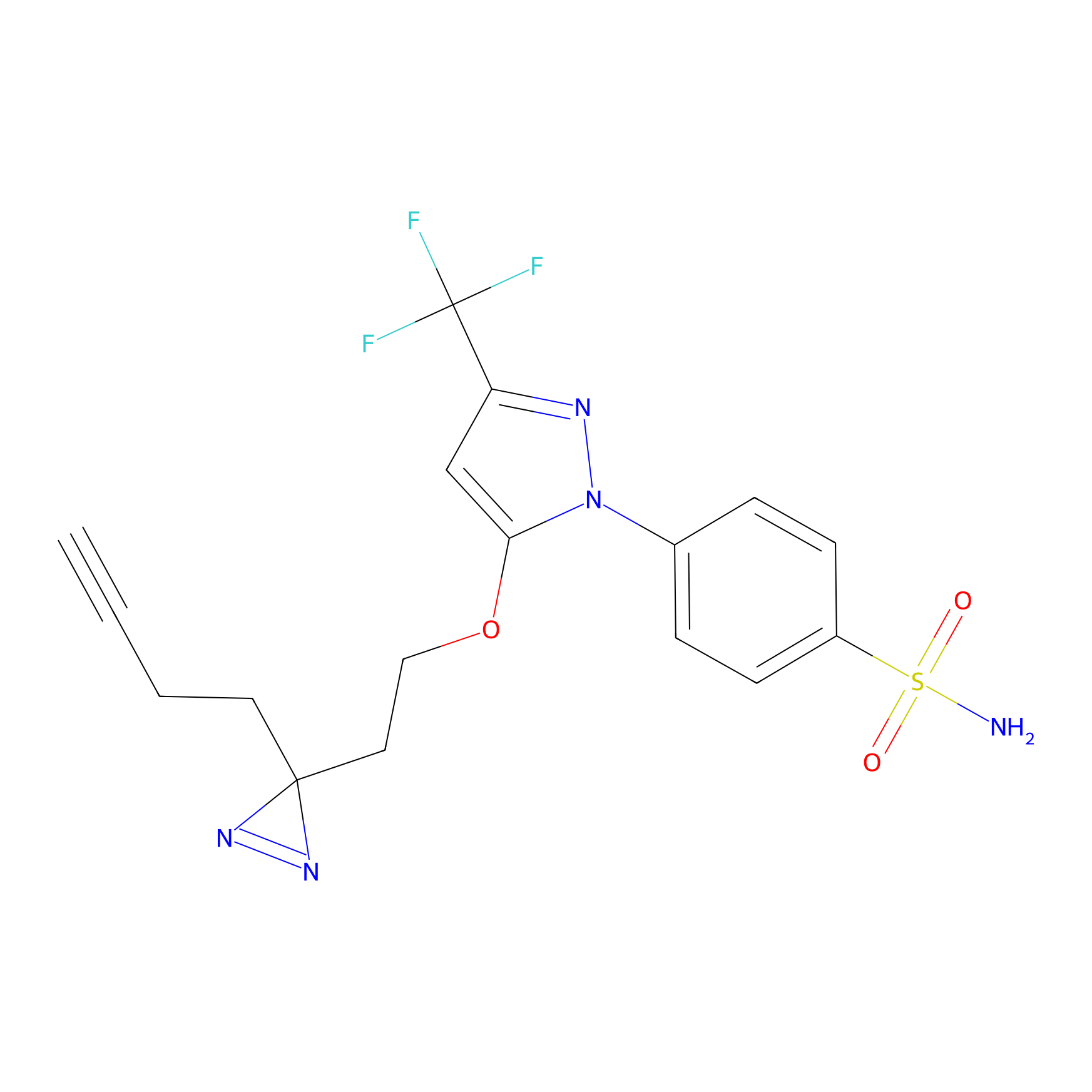 |
N.A. | LDD0019 | [9] | |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Fumarate hydratase, mitochondrial (FH) | Class-II fumarase/aspartase family | P07954 | |||
Other
References
